Project Summary
Use of Shotgun Metagenomic NGS Combined with a Previously-developed Bioinformatics Pipeline vs. Traditional Culture/Screening Diagnostics to Evaluate Ecology of AMR and Diagnose Salmonella spp. in Cattle Feces Before and After Ceftiofur Metaphylaxis
- Principle Investigator(s):
- E. Doster1, P. Rovira2, N. R. Noyes1, B. A. Burgess3, X. Yang2, M. Weinroth2, L. Linke1, R. Magnuson1, K. Jones4, C. Boucher5, J. Ruiz5, P. S. Morley1, K. E. Belk2
- Institution(s):
- 1Department of Clinical Sciences, Colorado State University
- 2Department of Animal Sciences, Colorado State University
- 3Department of Population Health Sciences, Virginia Tech
- 4Department of Biochemistry and Molecular Genetics, University of Colorado-Denver
- 5Department of Computer Sciences, Colorado State University
- Completion Date:
- June 2015
Background
Antimicrobial resistance is a pressing public health concern, due to both present‐day associations with increased morbidity and mortality, as well as fears of a “post‐antibiotic” future in which many antimicrobials are no longer effective in treating common infections. With recent increased attention on antimicrobial resistance, antibiotic use practices in food production systems have come under closer scrutiny, particularly with respect to injudicious use of antimicrobials. In 2011, the National Animal Health Monitoring System (NAHMS) Beef Feedlot Survey identified the most commonly used class of antimicrobials is macrolides, and since the FDA classifies macrolides as “critically important” to human medicine, it is necessary to understand the potential effect of the use of macrolides in beef production on antimicrobial resistance. Past research suggests that a traditional culture‐based approach, in which one or several organisms are selected and interrogated for resistance, is inadequate to describe and fully understand the role that antimicrobial use plays in influencing the quantity and quality of resistance gene presence, transfer and expression. Shotgun metagenomics, facilitated by next‐generation sequencing machines, represents a novel and fundamentally different approach to investigate complex microbial communities by sequencing all the DNA present in a sample. As sequencing costs continue to drop, combining all aspects of food safety surveillance and monitoring into a single shotgun metagenomic sample is likely to become a feasible and cost‐effective alternative to multiple workflows for testing for multiple pathogens, residues and resistance genes.
The overarching goal of this study was to develop and validate a dual‐purpose metagenomics pipeline for pathogen detection and resistome analysis of cattle feces in the early part of the feeding period. One objective under this goal was to utilize shotgun metagenomics to understand the impact of metaphylactic macrolide (tulathromycin) exposure on the resistome of cattle in the early feeding period. The second objective was to develop an approach for identifying Salmonella enterica in shotgun metagenomic data and to compare this approach to culture‐based results.
Methodology
A convenience sample of two pens in a Texas feedlot were selected for study enrollment. One pen was chosen to receive the antimicrobial intervention, while the other was chosen for the control. The only difference between groups was the additional administration of 8 cc per animal of the antimicrobial tulathromycin (Draxxin) to animals in the treated group by subcutaneous injection in the neck, upon arrival processing. Animals in both groups were processed according to standard feedlot procedures including vaccination, parasite control and application of growth implants. All animals received in‐feed Tylosin (a macrolide antimicrobial to prevent liver abscesses), at a target intake of 90 mg per head per day. Cattle were sampled at arrival processing and 11 days into the feeding period. Individual fecal samples (>25 grams) from the recto‐anal junction were collected, placed within sterile plastic bags, packed in coolers with frozen icepacks at 4° C and transported to the laboratory for processing within 8 hours of sample collection. Fecal samples were subjected to aerobic culture for Salmonella enterica identification and then frozen at ‐80°C prior to DNA extraction.
A subset of the study population, 15 cattle per group, were selected for DNA extraction and subsequent shotgun metagenomic sequencing. Corresponding fecal samples from both time points (n = 60) were thawed, 250 mg were collected, and whole community DNA was extracted using the Mo‐Bio PowerFecal DNA isolation kit, purified and subjected to shotgun sequencing on the Illumina HiSeq 2000. Sequenced reads were trimmed for poor quality and filtered to remove any bovine DNA present in the samples. Kraken, a taxonomic sequence classifier based on National Center for Biotechnology Information’s RefSeq database, was used to classify reads belonging to Salmonella enterica spp. and the software was subjected to custom synthetic metagenomic samples to test its accuracy. To evaluate the resistome, sample reads were aligned to a custom database of publicly available antimicrobial resistance genes, and only genes with >80% gene fraction were considered present for subsequent analysis. Descriptive analysis was performed on results and differences in resistome composition were explored using Nonmetric Multidimensional Scaling (NMDS) ordination and normalization with cumulative sum scaling. Differences in antimicrobial resistance gene abundance were assessed using a zero‐inflated Gaussian multivariable mixture model.
Findings
In general, aerobic culture suggests low Salmonella spp. prevalence (4/409) in our study
population and serogroup identification coincides with previous literature describing
serogroups C1 and K as commonly present in feedlot cattle operations. Polymerase chain
reaction results were negative for all 28 arrival processing samples that were tested using
this method.
Kraken identified Salmonella DNA from more than 10 serogroups in all 60 samples, but our testing disputes the accuracy with which Kraken is able to classify Salmonella enterica DNA down to the serovar level. Based on testing results, kraken‐based presence/absence and
within‐sample relative abundance of specific Salmonella enterica spp. enterica serovars
was determined to be unreliable when using the current reference RefSeq database, which
does not include many serovars commonly found in beef cattle, including the serogroup K
that was identified in the culture results in this study.
Across all 60 samples, it was identified that approximately 3.8 million reads aligned
to 343 unique resistance genes. Of these 3.8 million reads, 96.14% of these reads aligned
to genes that provide resistance to antimicrobial drugs, 1.68% to genes that provide
resistance to metals, and 0.88% to genes that provide resistance to biocides. Of the total
reads, 1.30% aligned to genes that provide resistance to a combination of antimicrobials,
metal and/or biocides (“cross‐category resistance”). Resistance richness and Shannon’s
diversity at the class, mechanism and gene levels were not significantly different between the
two treatment groups at either arrival processing or 11 days into the feeding period. Notably,
the abundance of macrolide and tetracycline resistance determinants (the two most abundant
classes of resistance) was not significantly different between the two groups at either
sampling time. Comparison of differentially abundant classes and mechanisms showed
increased abundance in the control group compared to the treated group in most cases.
However, most of these mechanisms were present at such low abundance that estimates of
log‐fold change between the treated and control groups are unreliable and/or not
biologically meaningful, and therefore these results should not be over‐interpreted.
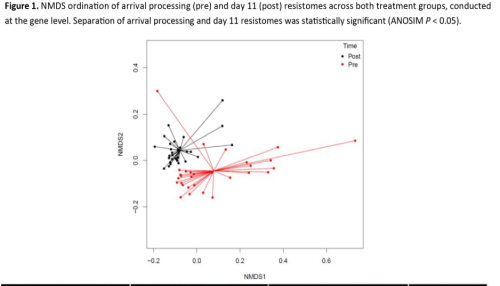
Interestingly, NMDS ordination (Figure 1) shows significant separation of the arrival processing and day 11 resistomes across both groups. This shift was observed at the gene, mechanism and class levels. Analysis of antimicrobial resistance determinant type and quantity suggests that the transition into the feedlot environment exerts a very strong and broad effect on the composition of the resistome; potential drivers of this effect are numerous, and include sudden co‐location of cattle from diverse locations, stress, a change in diet, vaccinations and in‐feed ionophores and antimicrobials. In this context, exposure to tulathromycin during arrival processing exerts a relatively small and narrow effect, which is most clearly seen in the significant decrease in resistome diversity among treated cattle, and the lack of such a change in control cattle. From a population genetics perspective, such decreased diversity of microbial genomes typically stems from fairly strong selective pressure acting on the microbial community. Therefore, this decrease may be the result of the additional antimicrobial exposures in the treated cattle. These results should also be considered in light of our previous metagenomic investigation into beef system resistome dynamics, which found a significant decrease in resistome diversity from the beginning to the end of the feeding period. Together, the results of these two studies suggest that selective pressure on the resistome may results from both duration and dosage of exposure to antimicrobials, i.e., exposure to tulathromycin at arrival processing result in a significant decrease in diversity, while short‐term exposure to Tylosin does not, but long‐term exposure to in‐feed antibiotics during the feeding period results in a reduced diversity resistome.
Implications
Although metagenomics provides a wealth of information, it appears that additional diagnostic techniques and/or more comprehensive genome databases are required to strengthen the interpretation of pathogen detection results and validate resistome analysis. Researchers surmise that as sequencing costs decrease, reference databases improve and bioinformatics pipelines are streamlined for easier use, the adoption of metagenomics will inevitably fit a complementary role in pathogen detection and surveillance as well as resistome analysis in the food industry.
Finally, the main challenge in adopting shotgun metagenomic methods becomes not only to analyze the data, but also to interpret the results accurately and engage in public discussion to provide biological context. In this particular instance, identifying DNA from Salmonella enterica in all of this study’s samples could be misconstrued to signify an overabundance of foodborne pathogens in beef production systems. However, with the added context that these results do not confirm the presence of live Salmonella enterica, and that bacterial culture results suggest low prevalence, any potential sensationalist claims are mitigated and public confidence on food safety preserved. Scientists and industry partners need to spearhead efforts to employ metagenomics in food production systems and develop a better understanding of the biological relevance of those results. With the new perspective into microbial ecology allowed by metagenomics, the livestock industry can revolutionize the study of food safety and livestock production to further bolster the food supply chain.