Project Summary
Use of Bovine‐specific Peptide Arrays for Kinome Analysis to Advance Understanding of Salmonella in the Peripheral Lymph Nodes
- Principle Investigator(s):
- R. J. Arsenault, T. R. Brown, T. S. Edrington
- Institution(s):
- USDA‐ARS, Food and Feed Safety Research Unit
- Completion Date:
- June 2015
Background
Species‐specific kinome peptide arrays designed for bovine biology have been successfully used in a number of contexts, including the study of Johne’s Disease and prion induced bovine spongiform encephalopathy (mad cow disease). The overlying principle involves designing a peptide array of kinase target sites, specifically designed to match the amino acid sequence in the species of interest. The data provides a valuable window into animal biology. Salmonella is a common gastrointestinal resident in both dairy and feedlot cattle in the Southwestern United States and more recently, it has been reported that the peripheral lymph nodes (PLN) of cattle often harbor Salmonella. Utilizing a relatively large number of PLN samples collected from other projects representing cattle slaughtered in the Southwestern United States, multiple, biologically relevant comparisons were conducted using kinome peptide arrays.
The objective of this study was to compare (using species‐specific kinome peptide arrays) the following:
- Subiliac lymph nodes from feedlot cattle that were infected or not infected with Salmonella;
- Subiliac lymph nodes from feedlot cattle harvested in either the summer or winter;
- Subiliac lymph nodes from feedlot cattle and cull dairy cows; and
- Lymph nodes from cattle experimentally inoculated with Salmonella versus naturally infected animals. Relative differences or similarities from these comparisons may illuminate the most biologically relevant differences between these animals in terms of animal health and susceptibility to disease.
Methodology
Subiliac lymph nodes were collected from cattle at slaughter. A small portion (approx. 2
g) of each node was removed and frozen at ‐80°C for later analysis. Tissue samples were
weighed, homogenized and subjected to the peptide array protocol. Data normalization,
statistics and clustering analysis were performed for the peptide using the Platform for
Intelligent, Integrated Kinome Analysis version 2 (PIIKA2) online software platform.
Geneontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis were performed by uploading the statistically significant peptide lists to the
Search Tool for the Retrieval of Interacting Genes (STRING).
Findings
The various analyses and group comparisons conducted herein present a broad overview of the power of kinome analysis, animal kinotyping and peptide phosphorylation comparative analysis. In comparing PLN that are Salmonella positive to those that are culture negative, it was demonstrated that individual animals display different kinome profiles centered on innate and adaptive immune responses to Salmonella. These cellular signal transduction changes brought about due to bacterial infection of the lymph nodes has never been considered at an active protein level.
In comparing feedlot cattle to cull dairy cattle, the data illustrated that the Salmonella infection state was more important than cattle breed or farm of origin. Also, it was the Salmonella infection and subsequent response that separated these two cattle types, as in their non‐infected (Salmonella negative) state, they looked remarkably similar at the kinome level. Understanding this separation following infection may help the beef industry understand why these two cattle types have a differential susceptibility to Salmonella acquisition by the PLN.
In contrast to the feedlot and dairy cattle comparison, the summer/winter lymph node comparison appeared to be defined by season more than infection status. Animals sampled in the winter, regardless of Salmonella status, were more alike than cattle sampled in the summer, which were widely variable in their response. This summer variation also appeared independent of Salmonella concentration within the lymph node. Using these same PLN, a representative summer and winter cattle kinotype was generated to show why different seasons appear to affect susceptibility to disease, with results suggesting that a significant immune component is involved.
Finally, lymph nodes from experimentally inoculated cattle were distinct in their kinotype
as compared to naturally infected animals. Further research into these differences is
warranted. These results could have significant implications for animal disease
experimentation, as much of this research is conducted on experimentally infected
animals and then applied to naturally infected animals. Further analysis of this data should
increase our understanding of these differences, so that future research utilizing experimentally infected animals will be properly analyzed and interpreted.
Implications
To the knowledge of the researchers of this study, this is the first research reporting the use of peptide kinome arrays in an attempt to increase the understanding of Salmonella in the PLN of cattle. Results of the comparisons suggest that differences in the immune system’s response to Salmonella may explain the differences in Salmonella prevalence in the PLN among fed and cull cattle. Additionally, it appears that season (summer in particular) and its influence on immune function, and not Salmonella status (infected versus non‐infected), may explain the seasonality of this pathogen in bovine lymph nodes. Furthering understanding of immune function in relation to cattle type and season may aid in the design and implementation of successful pre‐harvest intervention strategies.
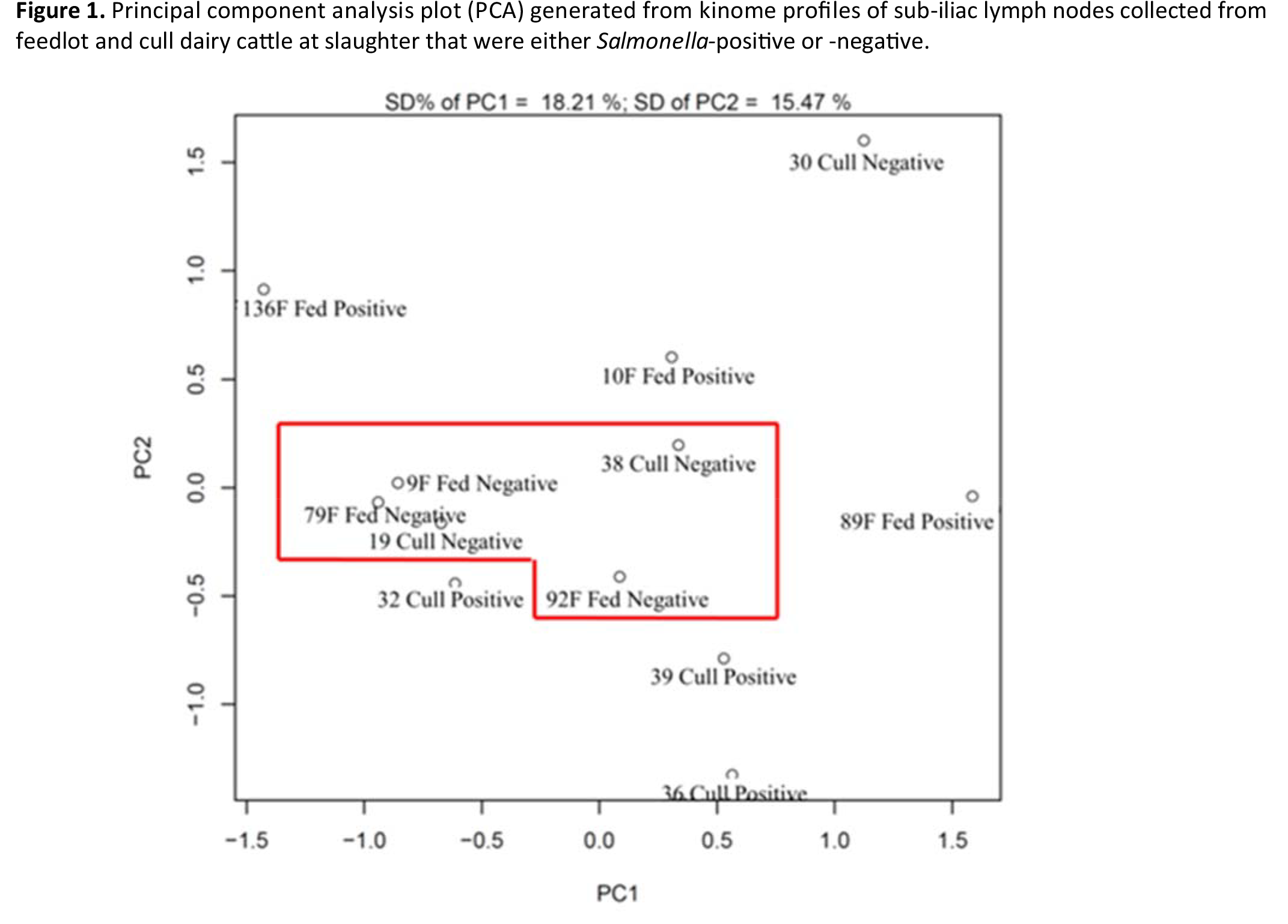
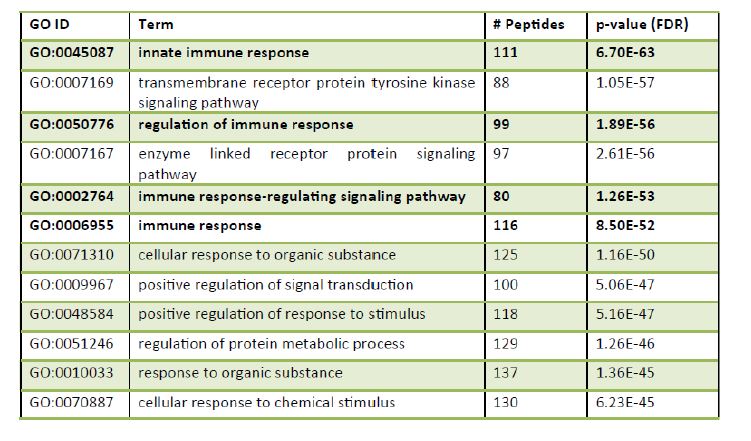
Table 1. Top 12 Geneontology (GO) biological processes in the sub‐iliac lymph nodes collected from feedlot cattle in the summer and winter months. A list of representative GO terms was generated by taking the statistically significant peptides from the comparison and inputting this peptide list into the online Search Tool for the Retrieval of Interacting Genes (STRING) database. Presented below are the top 12 terms from the input data. The pathways in bold are those involved in the immune response (total immune peptides 406).