Project Summary
Determination of Salmonella Lymph Node Contamination of Colonized and Naïve Cattle Post‐Comingling During the Summer in a Salmonella‐endemic Feedlot
- Principle Investigator(s):
- T. Arthur and D. Harhay
- Institution(s):
- USDA, ARS, Roman L. Hruska U.S. Meat Animal Research Center
- Completion Date:
- June 2015
Background
In spite of substantial efforts and investments, Salmonella harbored in cattle lymph nodes continues to present a major challenge to the beef industry. Researchers continue attempts to identify pre‐harvest interventions that are effective against Salmonella. However, until a clear route of infection is identified, pre‐harvest interventions are somewhat futile. Recent studies have shown a regional effect, where Salmonella appears to be more common and present in higher numbers in cattle environments found in the Southern United States. The cause of this regional effect is unknown at this time. It has been hypothesized it may simply be a function of temperature. However, recent findings have challenged that hypothesis. Preliminary data was collected for an investigation where a group of cattle, that had limited Salmonella exposure, if any, were transferred to a feedlot known to harbor Salmonella. In that study, the cattle were found to shed Salmonella in their feces, and all animals carried Salmonella on their hides. However, at slaughter, Salmonella was not found in the lymph nodes of any of the cattle sampled. While preliminary, these data suggested that management practices and exposures occurring prior to placement in the feedlot may affect lymph node colonization.
The objective of this experiment was to determine the prevalence and levels of Salmonella
in lymph nodes of naïve cattle and an equal number of cohort animals when comingled in
a feedlot that harbors Salmonella.
Methodology
Feeder cattle (70 head termed naïve for this study) were transferred from a BIFSCo
Region 5 feedlot to a BIFSCo Region 3 feedlot (Figure 1). Detection of Salmonella in the
Region 5 feedlot has been rare despite in‐depth sampling over several years. Conversely,
Salmonella has been commonly found in the Region 3 feedlot in several recent surveys.
The cattle from the Region 5 feedlot were termed “naïve” as they would have had little, if
any, interaction with Salmonella based on previous studies. Following transfer to the
Region 3 feedlot, the naïve cattle were maintained as a lot until re‐implant when they were
comingled with cohort cattle upon sorting into three different groups based on weight.
The cohort cattle came from BIFSCo Region 7. Sorted cattle remained in their designated pens until load out for slaughter.
Fecal and hide swabs were collected at the Region 5 feedlot for all animals to be shipped.
Sampling occurred prior to shipping. Fecal swabs and hide swabs were collected again
from the naïve cattle one day after arrival at the Region 3 feedlot, which was one week
after the exit samples were collected. Fecal swabs and hide swabs were collected from
the naïve cattle and an equal number of cohorts at sorting during re‐implant. At slaughter, fecal swabs and both sub‐iliac lymph nodes were collected from the naïve cattle and an
equal number of cohorts (Figure 2). For twelve head from each of the naïve and cohort groups, an addition four lymph nodes (two superficial cervical and two popliteal lymph nodes) were collected from each carcass. All samples were analyzed for Salmonella concentrations and prevalence.
Findings AND IMPLICATIONS
The prevalence for carcasses with at least one lymph node positive was not different
between naïve and cohort groups (Table 1). The results of this study show that naïve
cattle are susceptible to lymph node colonization by Salmonella. However, the prevalence
of animals with lymph nodes colonized by Salmonella was quite a bit lower than what has
been observed in other studies from this feed yard despite having similar hide and fecal
prevalence of Salmonella. The low prevalence observed in the current study could be due
to harvesting these cattle in November and December, when lower temperatures may
have caused a decrease in the environmental Salmonella load. Alternatively, it is possible
that not being exposed to Salmonella early in life may have predisposed the naïve cattle to
see Salmonella as foreign and be more resistant to lymph node colonization.
This study also showed that Salmonella prevalence in feces and especially on hides can
undergo dramatic shifts in very short periods of time due to the environmental presence of
Salmonella. Salmonella prevalence on the cattle hides rose from 1.4% to 97.1% with only 24
hours exposure to a high Salmonella environment. In addition, 96% (67 of 70) naïve cattle
at re‐implant had enumerable concentrations on their hides, which averaged 3.6 log CFU of
Salmonella/100 cm2 (Table 2). These results demonstrate the need to develop pen surface
interventions to reduce the environmental exposure to cattle entering a feedlot setting.
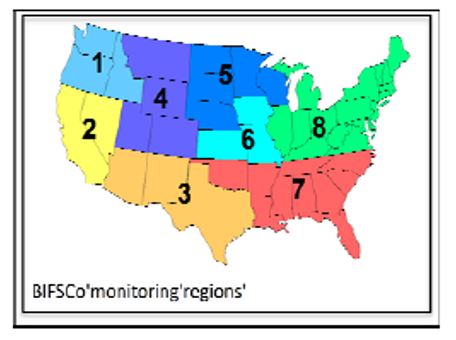
Figure 1. BIFSCo monitoring regions.
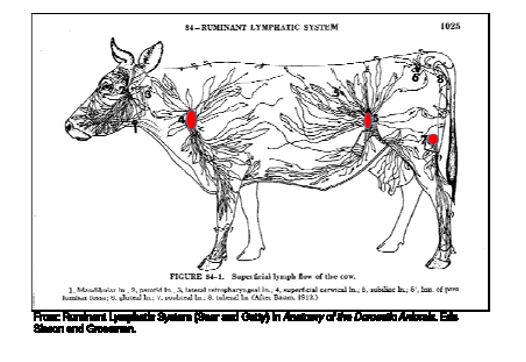
Figure 2. Ruminant lymphatic system anatomy.
Table 1. Salmonella prevalence.
|
Sample Period |
Sample Type |
Naïve |
Cohort |
|
Load out (May) |
Fecal swab |
0.0% |
- |
|
|
Hide swab |
1.4% |
- |
|
Incoming (May) |
Fecal swab |
15.7% |
- |
|
|
Hide swab |
97.1% |
- |
|
Re-implant (Sept.) |
Fecal swab |
75.0% |
85.7% |
|
|
Hide swab |
100% |
100% |
|
Harvest (Nov./Dec.) |
Fecal swab |
63.8% |
68.3% |
|
|
Lymph node (sub-iliac)a |
10.6% |
15.9% |
|
|
Lymph node (any)b |
14.9% |
15.9% |
-
aPrevalence of carcasses with at least one sub-iliac lymph node containing Salmonella.
- bAn additional four lymph nodes (2 superficial cervical and 2 popliteal) were collected from twelve of the naïve and twelve of the cohort carcasses. These data represent the prevalence of carcasses with any lymph node positive for Salmonella.
Table 2. Enumeration of Salmonella on the hides of naïve cattle.
|
Sample Period |
# of Samples with Enumerable Salmonella Concentrations |
Average log of CFU of Salmonella/100 cm2 |
|
Load out |
0 |
NA |
|
Incoming |
29 |
2.5 |
|
Reimplant |
67 |
3.5 |