Project Summary
Use of Metagenomic High‐throughput Sequencing Technology and Robust Bioinformatics to Assess the Microbiome of Cattle, their Environments, and Beef Products to Determine the Degree of Antimicrobial Resistance
- Principle Investigator(s):
- K. E. Belk1, D. R. Woerner1, N. R. Noyes1, P. S. Morley2, I. Geornaras2
- Institution(s):
- 1Department of Animal Sciences, College of Agricultural Sciences, Colorado State University
- 2Department of Clinical Sciences, College of Veterinary Medicine & Biomedical Sciences, Colorado State University
- Completion Date:
- May 2014
Background
Antimicrobial resistance (AMR) is a pressing public health concern, and the use of antimicrobials in food animals has been posited as a major driver of increasing AMR. However, studies in this area have failed to provide reproducible, actionable results, largely due to the limitations of culture‐based microbiological methods that necessitate reliance on “indicator” bacteria to represent the complex ecology of 1,400+ microbial taxa present in the bovine gut. Shotgun metagenomics enables a more valid approach, and the goal of this project was to use such an approach to trace AMR during beef production.
The objectives of this study were to use sophisticated molecular and statistical analysis methods to study evolutionary relationships and transmission of antimicrobial resistance genes among groups of organisms (microbiota) associated with cattle, beef, humans and the environments in which they cohabitate; and to determine the occurrence and abundance of microbes and genes in the cattle and beef product ecosystems by studying the microbiome of the beef production chain, with emphasis on the dissemination of antimicrobial resistance genetics.
Methodology
Research funded by the Beef Checkoff was conducted to determine prevalence and abundance of antimicrobial resistance genes in the cattle feeding and beef processing microbiome. Samples of feces, water and soil from two pens at four feedlots (one sample of each per pen per feedlot) that were located in two major, geographically dispersed cattle feeding areas were aseptically collected for high‐throughput complete DNA sequencing. Pen samples were collected at the time of cattle placement in the feedlot and again at the time that the same cattle were shipped for harvest. Subsequently, the same cattle were transported as market‐ready cattle to two differing packing plants (4 pens to each plant located in differing states) for harvest. Upon arrival at the packing plant, the trucks and cattle were identified and swab samples of trailer walls, samples of pen feces and water (two samples of each type per feedlot per plant) were collected aseptically for sequencing. Eventually, at the end of the production lines where products from carcasses of the original cattle were packaged and boxed, pooled swab samples from the trimming belt, round and chuck table as well as 400g of trimming samples (two sample of each type per feedlot per plant) were collected aseptically for DNA sequencing. Whole community DNA in each sample was extracted using Mo‐Bio PowerFecal® and PowerSoil® DNA Isolation Kit, purified, and subjected to metagenomics shotgun sequencing on Illumina HiSeq 2000. Sequence reads were trimmed and filtered before being assembled into contigs using SPAdes or IDBA‐UD. Contigs were compared to a database of AMR genes (ARG‐ANNOT) using BLAT. Only AMR genes with >80% coverage were considered present in the sample. Kraken, a taxonomic sequence classifier, was used to develop a phylogenetic profile for each sample.
Descriptive analysis was performed on results, and differences in AMR gene abundance between sampling locations were assessed using a zero‐inflated Gaussian multivariable mixture model.
Findings
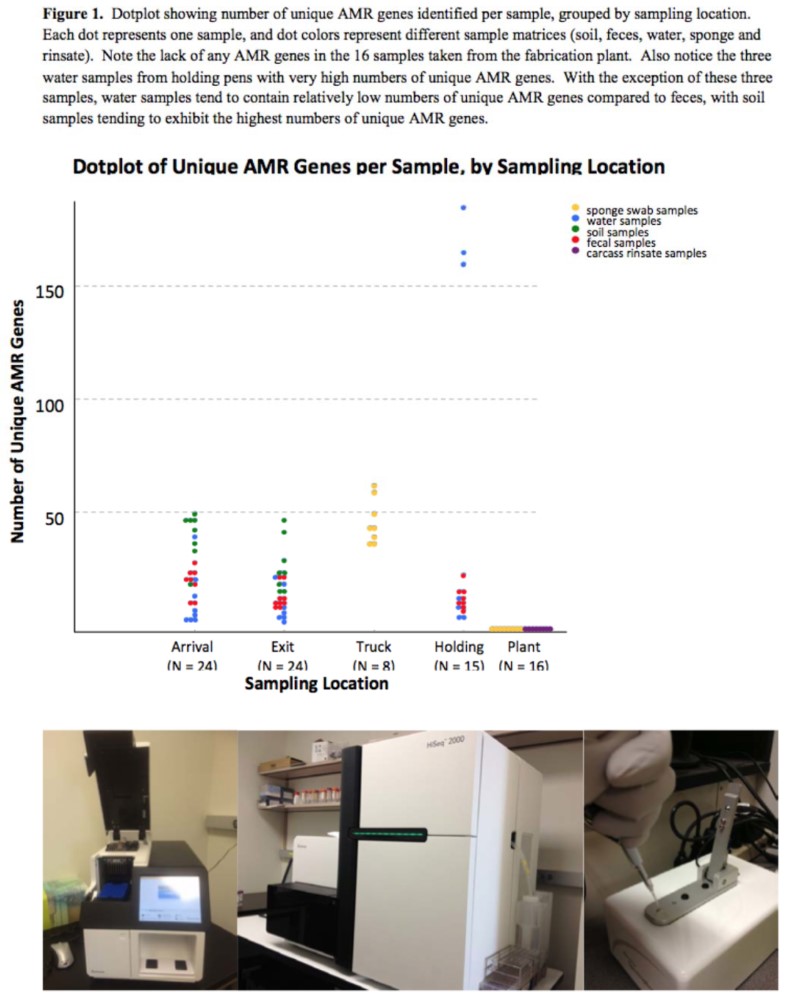
A total of 407.7 Gigabases of raw sequence data in 4.04 billion reads was produced. Mean Phred score was 35.2. Additionally, 90% of the bases in the reads had a quality score above Q30. The average sequencing reads per type of samples (arrival, exit, truck, holding pens, fabrication room) was 48M (ranging from 24 to 69M), 38M (ranging from 13 to 63M), 70M (ranging from 59 to 93M), 27M (ranging from 8 to 54M) and 60M (ranging from 47 to 85M), respectively. An average of 5.1% of reads was removed due to low quality. Assembly produced a mean of 533,157 contigs per sample, 10% of which were longer than 500bp (average per sample). In total, 266 unique AMR genes were identified when DNA from 87 samples was compared to the ARG‐ANNOT database of AMR genes. The median number of unique AMR genes identified per sample was 20, but the distribution between samples was highly skewed with significant changes between sampling locations. Transport truck samples exhibited the highest AMR burden with a median of 43.5 unique AMR genes per sample; however, none of the samples (n = 16) collected from the slaughter plant (belt swabs or carcass rinsates) harbored any AMR genes. In addition, three water samples taken from holding pens had very high numbers of unique AMR genes (155, 160 and 179), primarily due to the presence of many unique bla(TEM) genes.
Implications
Results of this project suggest that currently utilized slaughter‐based intervention strategies may serve dual‐purpose to reduce both pathogen load as well as AMR burden on meat products. Transport trucks may be a critical control point for containment of AMR determinants within the feedlot. Finally, the findings of this research support the use of metagenomics in gaining a more comprehensive understanding of AMR ecology in food production systems.