Project Summary
Identifying Bacteriophages Capable of Inactivating Multi‐drug Resistant (MDR) Salmonella Newport as a Pre‐harvest Antimicrobial Intervention
- Principle Investigator(s):
- T. Matthew Taylor, Kerri B. Gehring, Jason J. Gill, Jeffrey W. Savell, Yicheng Xie
- Institution(s):
- Texas A&M AgriLife Research; Texas A&M University
- Completion Date:
- May 2014
Background
The carriage of Salmonella (multidrug‐resistant or antibiotic susceptible) in the lymph system of cattle may contribute to pathogen transmission to consumers via ground beef and may cause foodborne disease if beef is improperly handled during its preparation. The U.S. beef industry has adopted a number of pre‐harvest antimicrobial interventions to reduce the entry of the non‐typhoidal Salmonellae into the post‐harvest environment. Bacteriophage (phage) are viruses that infect bacteria and are highly successful predators of bacteria. Phage have been shown to be effective in controlling a wide variety of pathogens in livestock species, including Salmonella in poultry and E. coli O157:H7 in cattle and sheep.
The objectives of this research were to: i) sample beef feedyards for the identification of Salmonella Newport‐specific bacteriophage, and; ii) characterize recovered phage for the ability to infect and lyse S. Newport and other serovars of S. enterica as a first step to the design of a novel phage‐based antimicrobial intervention strategy to prevent Salmonella carriage in the lymphoidal tissues of beef cattle.
Methodology
Three beef cattle feedyards located in south Texas were selected for sampling and subsequent Salmonella and phage isolation. Feedyards were chosen from amongst a set of cattle feedyards previously identified to be consistently positive or negative for Salmonella carriage in lymph nodes of cattle presented for harvest. Samples of dropped feces, animal pen soils, feed from feed bunks, and water from troughs were aseptically collected. Procedures for the selective enrichment and isolation of Salmonella from collected samples were adapted from standard methods. A set of 31 Salmonella isolates was selected for serotyping analysis to be representative of Salmonella‐positive feedyards and sample types. Feedyard samples were pooled by subsample to produce 36 composite samples containing the three subsamples collected from each pen and sample type. Samples were enriched for phage by a mixed‐host enrichment procedure. Phage were plated and cultured by the soft agar overlay method. For the isolation of individual phage, enrichments were pooled by pen. Individual plaques were picked from each plate, sub‐cultured and propagated by soft agar overlay.
Findings
The majority of Salmonella isolates were recovered from feedyard 1000, where all sample types yielded Salmonella isolates, though feces and soils samples yielded the greatest variety of Salmonella serovars. In the current study, 5.6% of feedyard 2000 samples were positive for Salmonella and all were from pen soil. Of the isolates submitted for serotyping, 19 were typed as S. Anatum, 4 were S. Montevideo, 4 were S. Muenchen, 2 were S. Altona, and there was one isolate each of S. Kentucky and Kralingen. All 36 samples contained phage active against at least one of the Salmonella hosts tested. Phage‐positive samples were distributed evenly over the three samples feedyard sites, suggesting that phage prevalence is not strongly correlated with culturable Salmonella. In the second round of phage detection using Salmonella isolates obtained from feedyard samples, phages were detected far less frequently, with only 12 of 36 samples yielding detectable phage upon enrichment. The detection of phage in samples appeared to be dependent on both host strain and sample type.
Implications
The identification of phage from environmental and fecal samples in feedyards, regardless of the recovery of viable Salmonella serovars, indicates an ecology exists in which host (Salmonella) and predator (phage) are interacting. If confirmed, such an ecology may be able to be harnessed for the development of novel Salmonella‐targeting pre‐harvest food safety interventions capable of reducing Salmonella prevalence on cattle feedyards and subsequent transmission of the pathogen to consumers via beef.

Figure 1. Feedyard sample collection for obtaining Salmonella and Salmonella-infecting phages.
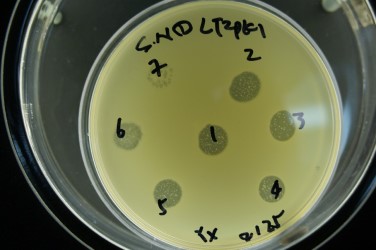
Figure 2. Petri dish with Salmonella host-infecting phages, indicated by presence of plaques (cleared zones) resulting from phage infection, lysis, and rupture of Salmonella cells with release of phage progeny into surrounding medium during incubation.
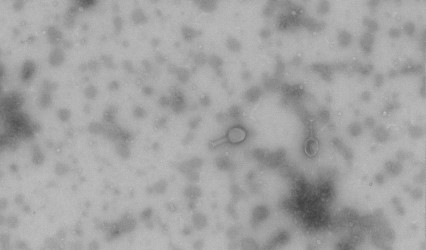
Figure 3. Transmission electron micrograph showing recovered phage and phage structure.
Table1 : Detection of Salmonella-infecting phage recovered from beef cattle feedyards and host ranges of isolated phage following enrichment using laboratory Salmonella serovars by feedyard, pen and sample type.
|
Sample |
Host Salmonella Serovara,b |
||||||||
|
Feedyard |
Pen |
Type |
Salmonella confirmed |
Typhimurium |
Typhimurium |
Newport |
Montevideo |
Anatum |
|
|
1000 |
A |
Feces |
+ |
+ |
+ |
+ |
- |
+ |
|
|
B |
Feces |
+ |
+ |
+ |
+ |
+ |
|||
|
C |
Feces |
+ |
+ |
+ |
+ |
+ |
- |
||
|
2000 |
A |
Feces |
+ |
+ |
+ |
+ |
+ |
||
|
B |
Feces |
+ |
+ |
+ |
+ |
- |
|||
|
C |
Feces |
+ |
+ |
+ |
+ |
+ |
|||
|
3000 |
A |
Feces |
+ |
+ |
+ |
+ |
- |
||
|
B |
Feces |
+ |
+ |
+ |
+ |
- |
|||
|
C |
Feces |
+ |
+ |
+ |
+ |
- |
|||
|
1000 |
A |
Feed |
+ |
+ |
- |
+ |
- |
- |
|
|
B |
Feed |
+ |
- |
+ |
- |
- |
|||
|
C |
Feed |
+ |
- |
+ |
- |
- |
|||
|
2000 |
A |
Feed |
+ |
- |
+ |
- |
- |
||
|
B |
Feed |
+ |
- |
- |
- |
- |
|||
|
C |
Feed |
+ |
- |
+ |
- |
- |
|||
|
3000 |
A |
Feed |
+ |
- |
+ |
- |
- |
||
|
B |
Feed |
+ |
- |
+ |
- |
- |
|||
|
C |
Feed |
+ |
- |
- |
- |
- |
|||
|
1000 |
A |
Soil |
+ |
+ |
+ |
+ |
+ |
+ |
|
|
B |
Soil |
+ |
+ |
+ |
+ |
+ |
+ |
||
|
C |
Soil |
+ |
+ |
+ |
+ |
- |
+ |
||
|
2000 |
A |
Soil |
+ |
+ |
+ |
+ |
+ |
||
|
B |
Soil |
+ |
+ |
+ |
+ |
- |
|||
|
C |
Soil |
+ |
+ |
+ |
+ |
+ |
+ |
||
|
3000 |
A |
Soil |
+ |
+ |
+ |
+ |
+ |
||
|
B |
Soil |
+ |
+ |
+ |
+ |
- |
|||
|
C |
Soil |
+ |
+ |
+ |
+ |
- |
|||
|
1000 |
A |
Water |
+ |
+ |
+ |
+ |
- |
- |
|
|
B |
Water |
+ |
+ |
+ |
- |
- |
|||
|
C |
Water |
+ |
+ |
+ |
+ |
- |
- |
||
|
2000 |
A |
Water |
+ |
+ |
+ |
- |
- |
||
|
B |
Water |
+ |
+ |
+ |
- |
- |
|||
|
C |
Water |
+ |
+ |
+ |
- |
- |
|||
|
3000 |
A |
Water |
+ |
+ |
+ |
- |
- |
||
|
|
B |
Water |
+ |
+ |
+ |
- |
- |
||
|
C |
Water |
+ |
+ |
+ |
- |
- |
|||
|
Total positive |
36 |
27 |
34 |
16 |
10 |
||||
aSalmonella host serovars were provided by the Food Microbiology Laboratory, Department of Animal Science, Texas A&M University (S. Typhimurium LT2), or by Tom Edrington, USDA-ARS, College Station, TX (S. Typhimurium USDA1, S. Newport USDA2, S. Montevideo USDA3, S. Anatum USDA4).
bPhage enrichments were conducted by adding 10 mL sample supernatant to 40 mL sterile TSB in 250 mL flask, and inoculation of each flask with 25 µL of overnight culture of each of the five enrichment Salmonella strains. Enrichments were incubated overnight at 37°C with aeration, 5 mL of culture was removed, centrifuged at 8,000 x g, 10 min, to pellet the remaining bacteria, and the phage-containing supernatant filter-sterilized through a 0.2 µm syringe filter. Sample enrichments were stored in the dark at 4°C until further use.