Project Summary
Discovery of Molecular Targets for Identifying the Top Six Human Disease-Causing Shiga-toxigenic E. coli non-O157 Strains
- Principle Investigator(s):
- James Bono and Gregory Harhay
- Institution(s):
- U.S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2012
Background
Over 100 different serotypes of Shiga-toxigenic E. coli have been reported to cause disease in humans. In North America, approximately half of human cases are caused by the serotype O157:H7, with the remaining cases being caused by non-O157 serotypes. From the non-O157 serotypes, six serotypes are responsible for 70-80% of the reported cases. Because of the number of reported cases of these six non-O157 serotypes and their ability to cause the same disease in humans as E. coli O157, the Food Safety Inspection Service has deemed the six non-O157 serotypes to be adulterants in beef trim. Twenty years of E. coli O157:H7 research has led to the development of reliable culture methods and molecular tests for detecting and identifying this pathogen. The molecular tests for E. coli O157 have been developed because genomic sequencing has given researchers a better understanding of the genome. However, little is known about the genomic content of other non-O157 STECs. In order to design molecular markers for non-O157 STEC serotypes, addition genomic sequence is needed for genome comparison. The more strains used when comparing genomes, the better chance of finding serotype specific molecular markers. Currently, there is genomic information in GenBank for one strain from three non-O157 STEC, O26:H11, O111:H8 and O103:H2. The researchers of this study sequenced the genomes of representative strains from six of the most reported non-O157 serotypes that have caused disease in humans (O26, O111, O103, O121, O45 and O145). This approach provided a more robust method for finding genomic variation that is distinct to each non-O157 STEC serotype and provide candidate molecular markers for assay development that distinguish STEC O26, O111, O103, O121, O45 and O145 serotypes.
The stated objectives for this work were to:
- Collect genomic sequences of 30 STEC non-O157 strains of the following serotypes; O26, O111, O103, O121, O45, and O145.
- Identify serotype specific DNA sequences that can be used to develop a molecular based assay for the detection of the top 6 human disease causing non-O157 STECs.
Genomic Sequencing of non-O157 STEC and Sequence Analysis
Five strains from non-O157 STEC serotypes O26, O45, O103, O111, O121, and O145 were sequenced using in-house integrated and automated microbial genome sequencing and annotation system that takes as input a microbial isolate and outputs an annotated assembled genomic sequence suitable for GenBank submission. Genomic DNA was extracted from each strain using a Qiagen DNA extraction kit, and shotgun DNA sequencing libraries were made for both the PacBio and 454 DNA sequencers. PacBio single molecule real-time sequencing (SMRT) reads were error corrected using the program pacBioToCa with 454 sequencing reads to generate draft genome assemblies. Contigs from the draft genomes were annotated using Do It Yourself Annotator (DIYA) running on an in-house computer to produce a GenBank ready file.
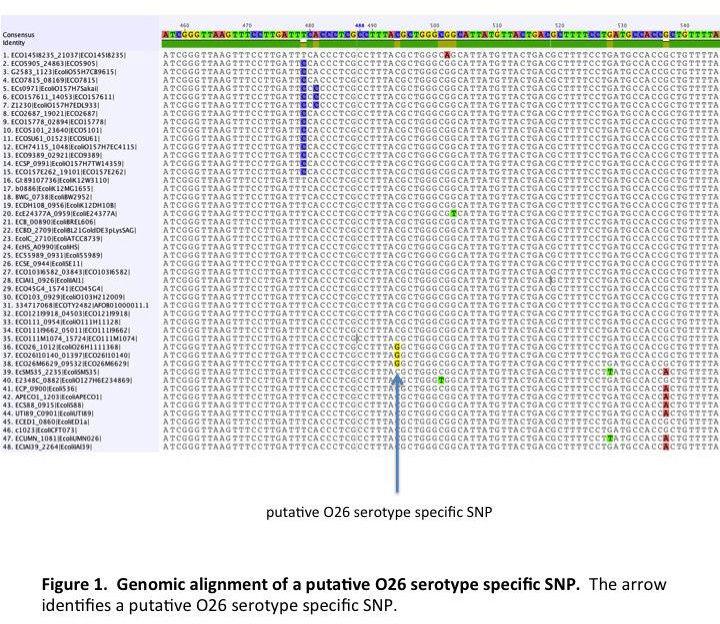
Genome Comparison and Nucleotide Polymorphism Validation
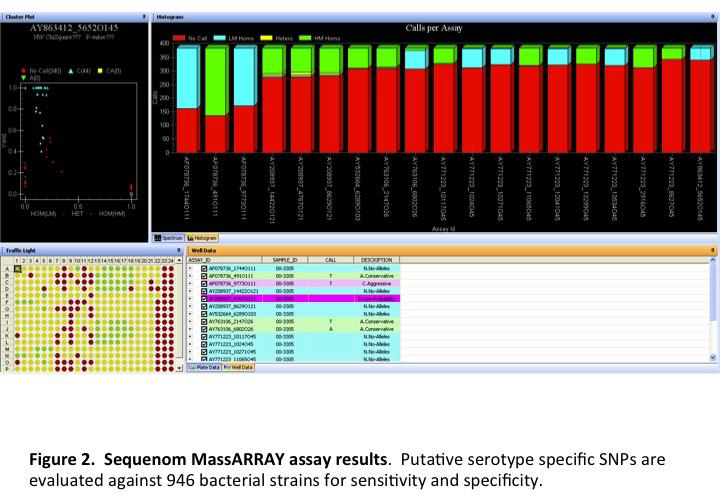
Annotated genomes were compared using software developed by current collaborators that is still under development. The software concatenates genes common to all the strains being compared. The concatenated sequences are aligned and used to build a tree based on relatedness. Nucleotide polymorphisms responsible for the branches, which are predicted to be unique to strains on that branch, were exported and used to design assays for the Sequenom MassARRAY analyzer. Depending upon the serotype, between 32 and 37 SNPs from serotype specific branches were assayed using the Sequenom MassArray analyzer. A total of 768 strains were used to validate the serotype specific SNPs and included one hundred ninety-two O157:H7, three O157:non-H7, four O55:H7, two O55:H6, eighty-three O111 STEC, twenty-three O111 non-STEC, eighty O26 STEC, thirty O26 non-STEC, nine O45 STEC, three O45 non-STEC, twenty-four O103 STEC, seven O103 non-STEC, five O145 STEC, one O145 non-STEC, six O121 STEC, six O121 non-STEC, eleven other E. coli STEC, one hundred seventy-six E. coli O-antigen standards, sixty-one Salmonella, and 42 other bacteria.
Findings
Serotype specific SNPs were identified for STEC O145:NM (n = 3), O121:H19 (n = 10) and O111:H8 (n = 2). No serotype specific SNPs were identified for STEC O26:H11, O103:H2, or O45:H2 because the proposed serotype specific SNPs were found in other bacterial strains.
Implications
The serotype specific SNPs identified for STECs O145:NM, O121:H19 and O111:H8 will allow quicker identification of samples that contain these foodborne pathogens. A material transfer agreement has been signed with a diagnostic company to use these serotype specific markers in a commercial assay. The anticipated outcome is a more accurate assay that will enable the end user to obtain results quicker, so perishable food products can be released, thus extending the shelf life of the product and saving the producer from lost product and increasing the safety of food for US consumers.
Table 1. Number of serotype specific SNPs identified by genome comparison.
|
Serotype |
Number SNPs |
Number Serotype Specific SNPs |
|
O26 |
36 |
0 |
|
O45 |
33 |
0 |
|
O103 |
33 |
0 |
|
O11:H8 |
33 |
2 |
|
O121:H19 |
33 |
10 |
|
O145:NM |
33 |
3 |