Project Summary
Characterization of Salmonella Gene Expression Profiles when Present in Cattle Subiliac Lymph Nosed – Proof of Concept
- Principle Investigator(s):
- Dayna M. Brichta-Harhay, Tim P. L. Smith, Jim Bono, Mick Bosilevac, Gregory P. Harhay
- Institution(s):
- U.S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2012
Background
Cattle are a noted reservoir for the foodborne pathogen Salmonella enterica, and ground beef has been identified as a possible mode of transmission for outbreaks of salmonellosis. Surveys of Salmonella contamination present in ground beef indicate that prevalence is generally low, and that when isolated the most commonly observed serotypes are Montevideo and Anatum. However, outbreaks attributed to ground beef are more often the result of contamination with Salmonella Newport or Typhimurium and rarely attributed to the more commonly observed serotypes. With the potential for impending regulation on the presence of particular Salmonella serovars contaminating meat, it is becoming more likely that beef processors will need tools for rapidly identifying certain Salmonella serotypes when detected.
The stated objectives for this work were to examine the differences in gene content and determine genetic markers (single nucleotide polymorphisms or SNPs) for the differentiation of serotypes more likely to be found in ground beef, namely Montevideo and Anatum, from those more likely to cause disease when present, namely Newport and Typhimurium.
Methodology
A diverse set of 48 strains of each stereotype were collected from cattle hide, carcass, fecal, and ground beef samples. Genomic DNA was isolated, pooled for each serotype, and then sequenced. Sequence reads were mapped to a Salmonella Newport reference genome and SNPs were determined. Those SNPs with the greatest coverage (highly represented in the DNA sequence and likely to be representative of the majority of strains present in the serotype pool) and with a variant frequency of greater than 80% (indicating that the majority of strains in the serotype pool shared that sequence) were cataloged and will be used for validation.
Findings
The data collected provided information on which SNPs can be used to differentiate the more medically relevant Salmonella (Newport and Typhimurium) from those that are potentially more commensal (Montevideo and Anatum).
Implications
Identifying molecular targets for differentiation of certain Salmonella serotypes will provide the beef industry with the tools needed to screen for particular Salmonella serotypes if present in ground beef or trim, thus reducing the chances of introducing these pathogens into the food chain and minimizing the potential for recalls associated with Salmonella contamination.
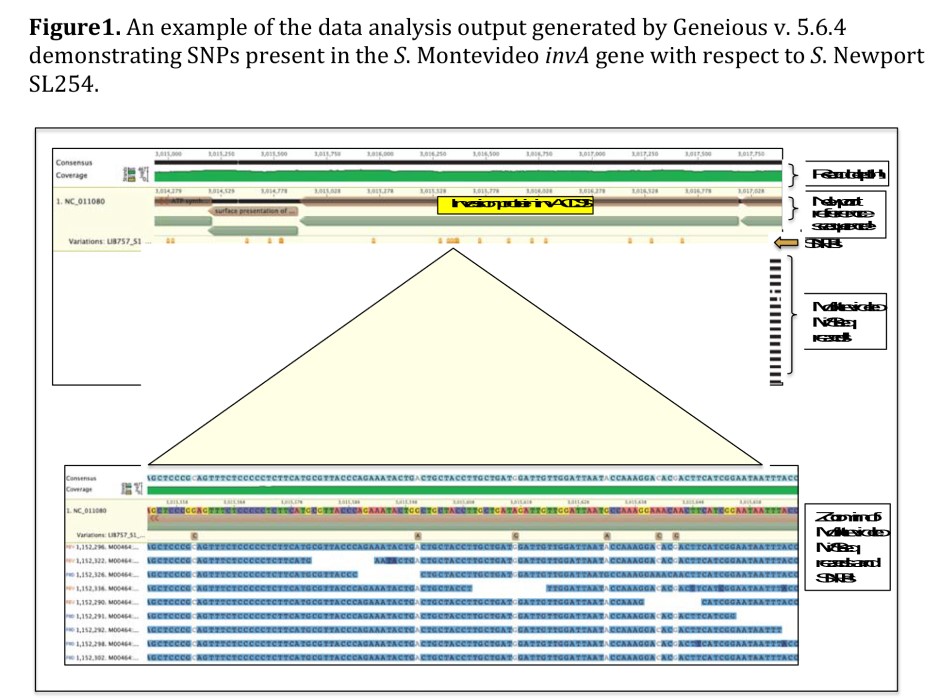
Figure 2. Salmonella growing on an XLD plate.
Figure 3. Electron micrograph of Salmonella enterica.