Project Summary
Characterization of E. coli O157:H7 Strains Associated with High Shedding Events
- Principle Investigator(s):
- Terrance M. Arthur, Rafiq Ahmed, Margo Chase-Topping, Norasak Kalchayanand, John Schmidt, James Bono
- Institution(s):
- U.S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2012
Background
E. coli O157:H7 is an important foodborne pathogen which poses a serious public health concern and financial burden. Cattle are the principal animal reservoir of E. coli O157:H7, and while rumen has been shown to harbor this pathogen on occasion, the rectal-anal junction (RAJ) has been shown to be the predominant colonization site. Once colonized, an animal can shed various amounts of E. coli O157:H7 in the feces. Super-shedders (RAJ colonized at greater than 104 CFU/g) have a significant effect on contamination of the cattle hide and carcass and are reported to be responsible for increased transmission of E. coli O157:H7 within production environments. Therefore, it is critical to identify, minimize or eliminate super-shedders from the cattle population in order to reduce E. coli O157:H7 transmission and beef carcass contamination for enhancing food safety.
Several approaches for reducing E. coli O157:H7 colonization of the cattle gastrointestinal tract (GIT) have been experimentally tested, such as different feeding regiments, feed additives, probiotics, and vaccines. To date, most of these studies have either been inconclusive, or treatments have only modestly affected colonization. More importantly, none of these interventions have tested whether the prevalence of super-shedding is reduced in cattle populations. This is significant, as modeling studies suggest that possibly as high as 96% of E. coli O157:H7 isolates originate from super-shedding animals. It is evident that a more thorough understanding of the factors promoting super-shedding is needed before we effective evidence-based methods of reducing transmission of STEC from cattle populations to the food supply can be designed. The super-shedding phenomenon can be broken down in to three principal components: (1) phylogenetic lineage of the colonizing O157:H7 strain, (2) the community composition of the microflora of the rectal-anal junction, and (3) the innate and adaptive immune response of the host. This proposal is designed to determine the contribution of strain type in super-shedding.
The stated objectives for this work were to:
- Identify cattle shedding E. coli O157:H7 at levels (> 104 CFU/g or swab).
- Isolate E. coli O157:H7 strains from super-shedders.
- Characterize strains to identify super-shedder specific traits.
Animals and Sample Collection
Fecal samples were collected from approximately 3,500 cattle at slaughter in commercial processing plants and 1,500 cattle in commercial feedlots. Swab samples were collected by swabbing the rectum, then placing the swabs in tubes containing broth to support bacterial growth. Tubes were stored on ice for shipment to the laboratory.
Isolation of E. coli O157:H7
Upon arrival at the lab, the sample tubes were mixed vigorously for 30 sec then samples were spread onto agar plates for enumeration of E. coli O157:H7. When detected, counts of O157:H7 were reported as colony forming units (CFU) per swab. Animals were classified as super-shedders when counts were greater than 104 CFU/swab. Up to twenty presumptive E. coli O157:H7 colonies were picked for confirmation.
Strain Characterization
All E. coli O157:H7 strains obtained from super-shedders were analyzed to identify strain-specific traits related to super-shedding. Genetic analyses were performed to determine strain lineages and other distinguishing features related to individual genotypes. Lineages of E. coli O157:H7 have been designated as I, II, and I/II. Lineage I/II share characteristics from both lineages I and II. While all three lineages have been found in cattle, only lineages I and I/II are typically associated with human disease.
Findings
Super-shedder prevalence among lots ranged from 0% to 4.5%. Overall, super-shedders were identified as 2.3% (n = 102) of the bovine population tested.
Up to twenty isolates were characterized per super-shedder sample. In every case, all isolates originating from a sample were identical in the genotypic and phenotypic characteristics tested. This indicates that within an animal, super-shedding is a function of a singular strain type.
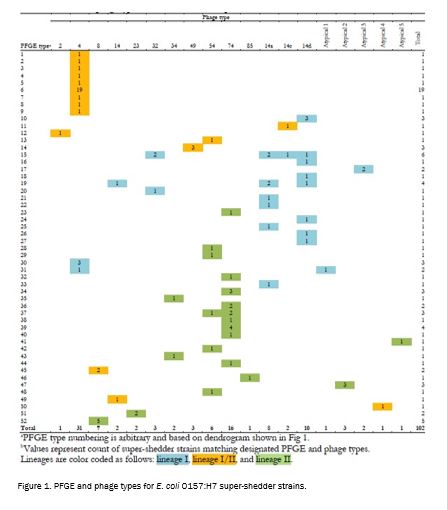
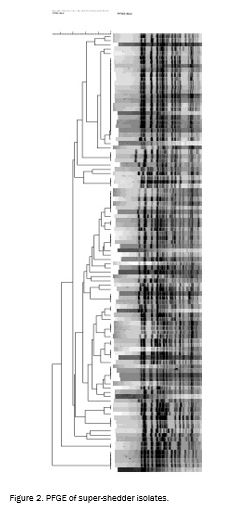
The super-shedder strains collected in this study were spread among the three lineages of E. coli O157:H7 with the majority (36%) of strains from lineage I/II and the minority (29%) from lineage I. PFGE types were not found to contain strains from multiple lineages. However, certain phage types did contain strains from multiple lineages. Super-shedder strains were not found to be restricted to particular PFGE or phage types.
Implications
In this study, no strain-specific components were identified as necessary for the development of super-shedding. Therefore, any E. coli O157:H7 can achieve the high cell levels associated with super-shedding. This data suggests that other super-shedding determinants, such as host genome/immune response and intestinal microbiome, should be investigated for the development of super-shedder intervention schemes.