Project Summary
Antibiotic Resistance by Source: Human‐Cattle‐Swine
- Principle Investigator(s):
- Terrance Arthur, Getahun Agga, John Schmidt, Tim Smith
- Institution(s):
- USDA, ARS, U.S. Meat Animal Research Center (USMARC)
- Completion Date:
- June 2014
Background
Clearly, antimicrobial resistance (AMR) predates the use of antimicrobials (AMs) by humans in clinical and agricultural settings. For the majority of AMs, the organism responsible for AM production would, by necessity, harbor a resistance mechanism to avoid its own termination. The problem arises when these resistance mechanisms are transferred to other bacterial species. The spread of AMR threatens the effectiveness of perhaps the most significant therapeutics available to maintain human health. Animal agriculture has been accused of encouraging the spread of resistance through the consumption of large quantities of antimicrobials for both therapeutic and prophylactic applications. Indeed, several studies have identified resistant bacterial strains in agricultural settings. The deficiency in these studies is that other environments were not sampled for comparison. When AMR is reported in agricultural settings without comparison to other environments there is a false pretense that the identified resistance is confined to the agricultural setting and would not be found elsewhere. The hypothesis for this project is that resistance elements will be found whenever and wherever they are looked for.
Many studies have identified populations of resistant microbes in a variety of habitats, ranging from confined animal feeding operations, to municipal waste streams, to pristine environments with little to no human impact. However, very little has been done to show that the habitat is not the issue, but rather that antibiotic resistance is a very widespread phenomenon.
The objective of this study was to determine the baseline prevalence of antimicrobial resistance in multiple environments.
Methodology
A total of 174 liquid and solid samples were collected from the effluent of three municipal sewage treatment facilities, three cattle feedlot runoff catchment ponds, three swine waste lagoons, and two environments not considered to be impacted by human or agricultural fecal waste. All sample sites were located in central and eastern portions of Nebraska.
For the wastewater treatment plants, liquid samples were collected at the location of discharge into the environment. The collection of solid samples varied by site, but all solid samples were obtained from material that was to be or had been released to the environment. Collection of solids samples at the cattle feedlots utilized manure storage piles if available, otherwise samples of pen surface material were collected. In swine production, solid waste is flushed from the production housing with the liquid waste, both flowing into a lagoon. As such, solids samples were collected around the edge of each lagoon.
Four samples of each sample type from each site were collected both in the summer and winter of 2013. Individual samples (n = 174) were processed by traditional culture techniques for AMR Gram negative (E. coli and Salmonella) and Gram-positive (Enterococci) bacteria to determine prevalence and to enumerate resistant strains. In addition, samples from each four samples of each sample type from each site were collected both in the summer and winter of 2013. Individual samples (n = 174) were processed by traditional culture techniques for AMR Gram-negative (E. coli and Salmonella) and Gram-positive (Enterococci) bacteria to determine prevalence and to enumerate resistant strains. In addition, samples from each of the following bacteria were the subjects of investigation in this project: 3rd‐generation cephalosporin‐resistant (3GCr) E. coli; folate synthesis inhibitor combination‐resistant (FSIr) E. coli; 3GCr Salmonella spp.; quinolone‐resistant (QNLr) Salmonella spp.; and macrolide‐resistant (MACr) Enterococcus spp.
Findings
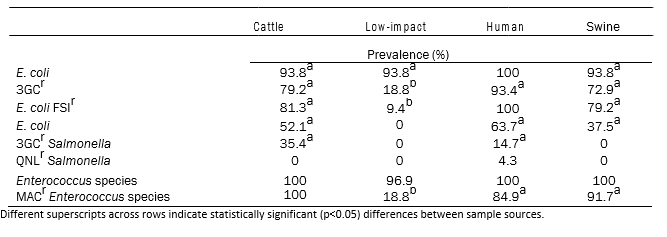
There were no statistically significant differences in the prevalences of 3GCr‐ and FSIr‐resistant E. coli obtained from cattle, human and swine waste samples (Table 1). Similarly, while the concentrations of 3GCr‐ and FSIr‐E. coli were higher in the municipal environment as compared to the cattle or swine environments, this difference was not statistically significant. 3GCr‐ and FSIr‐E. coli were commonly found in cattle, human and swine waste samples (all prevalences > 70%). 3GCr‐resistant Salmonella were recovered from only the cattle and human‐associated waste streams. Nalidixic acid resistant‐Salmonella were recovered from two samples collected at one municipal environment in the summer. Enterococci prevalence did not differ between any environments, while the prevalence of MACr‐Enterococci did not differ among cattle, human, and swine‐associated environments, but was significantly lower for low impact environments.
Implications
In this study it was demonstrated that AMR is a very widespread phenomenon, and that similar levels of ARB and ARG can be obtained from human, cattle, and swine waste. In addition, several ARG were detected in the low‐impact environment samples in spite of the fact that reservoir populations (generic E. coli and Enterococci) in those environments were low. This data shows that animal agriculture impacts the spread of AMR at the same level or possibly less than the materials released from municipal wastewater treatment plants.
Note for abbreviations: 3GCr = third generation cephalosporin resistant; FSIr = trimethoprim/sulfamethoxazole resistant; QNLr = quinalone resistant; MACr = macrolide resistance
Figure 2. Amplification curves from detection of ARG.
Table 1: Model adjusted prevalence of E. coli, Salmonella and Enterococcus spp. from cattle (n = 48), low‐impact environment (n = 32), human (n = 46) and swine (n = 48) samples.