Project Summary
A New Approach to Control E. coli O157 and Other Non‐O157:H7 Shiga‐toxin Producing E. coli (STEC) in Beef Cattle Using Natural Bacterial Competitors ‐ Phase 2
- Principle Investigator(s):
- N. D. Aluthge, Y. A. Wanniarachchi, C. J. Schneider, T. Klopfenstein, G. Erickson, J. Stratton, S. C. Fernando
- Institution(s):
- Department of Animal Science, University of Nebraska, Lincoln
- Completion Date:
- May 2013
Background
Cattle constitute a major natural reservoir of STEC, and cattle farming can be considered a major point at which intervention strategies can be targeted to minimize the prevalence of these pathogens. Reducing the load of pathogenic STEC at pre‐harvest can lead to significant improvements in the safety of beef throughout the food chain right up to the consumer. It has been shown that the prevalence of STEC among cattle in a pen varies widely. The exact reason(s) why certain animals within a pen shed higher numbers of STEC, whilst others that comingle with them within the same pen do not shed, is currently unknown. It is hypothesized that factors such as competition for nutrients and colonization space between STEC and native microbial communities within the intestinal tract of cattle may play a role in determining the animal’s shedding phenotype (high‐shedder or low‐shedder). This study aimed to characterize the microbial communities of STEC high‐shedding and low‐shedding cattle populations using high throughput genome‐sequencing technologies, to identify particular microbial populations, which may have a protective effect against the colonization of cattle by STEC and isolate candidate microbial species to use as direct fed microbials (DFM) to reduce STEC shedding.
The objectives of this study were to 1) isolate bacterial species into pure culture based on sequencing information and grow in large quantities; and 2) perform a feeding trial to evaluate the effectiveness of the identified direct fed microbial in reducing E. coli O157 and other non‐O157:H7 (Big Six) STEC shedding.
Methodology
During the months of August to September 2011, rectal grab samples of feces were collected weekly for 3 weeks from 170 animals housed at the ARDC Mead UNL research facility. Each of these fecal samples was subjected to microbiological analysis to detect and quantify Shiga toxin‐producing E. coli (STEC) levels in feces. Briefly, for each fecal sample, 10 g was diluted in 90 mL of phosphate‐buffered tryptic soy broth and was blended with a stomacher (200 rpm, 1 minute). From each fecal suspension, a 1 mL aliquot was removed, and 50 microliters was plated on the surface of a CHROMagarTM STEC plate using a spiral plater and incubated overnight at 42°C. Characteristic colonies (mauve color) were counted and the number of STEC/g of feces was calculated. Based on these results, 48 candidate STEC high‐shedding and low‐shedding animals were identified for high‐throughput sequencing.
The raw data from high‐throughput sequencing was processed through a quality filter and was analyzed using the analysis pipelines currently in place at the PI lab. The processed data was used for taxonomy‐based and phylogeny‐based analyses. Briefly, after removing low quality reads, the dataset was subsampled, and the phylogenetic diversity of the resulting reads were analyzed using tools available at the Ribosomal Database Project. In addition, the microbial communities were also analyzed using OUT‐based, analysis methods using “MOTHUR” analysis tools. ANOSIM and PERMANOVA analysis was carried out to ascertain whether there was a statistically significant difference between the microbial community compositions of the low‐shedder and high‐shedders. OTUs driving the difference between the two categories (i.e., high‐shedders and low‐shedders) were identified using ANOVA‐based test.
Based on the analysis of relative abundances of bacterial 16s rRNA reads between high‐shedders and low‐shedders, candidate microbial species were identified that were high in low‐shedders, that they may be important in outcompeting STEC by reducing their colonization and/or inhibiting their growth in the cattle gut environment. The selected candidate species were isolated from cattle feces using anaerobic culturing methods and PCR assays.
Findings
The fecal microbial community structure of STEC high‐shedding and low‐shedding animals are significantly different, suggesting that gut microbial community structure may influence the shedding phenotype. Microbial communities differentially present in low STEC‐shedding animals may be used as candidate microbial species for direct‐fed microbials to reduce STEC shedding. Currently, the candidate microbes isolated are being used in a feeding trial to estimate the applicability of using such microbes as direct‐fed microbials to reduce STEC shedding.
Implications
This new approach to control E. coli O157:H7 and other non‐O157:H7 will help make our ground beef safer and will help enhance the image of the industry. This research will help provide new methods to reduce the shedding of non‐O157:H7 in cattle and will help the industry to make strategic decisions in the future to make beef safer.
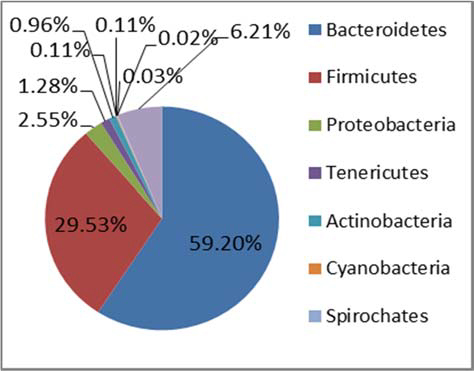
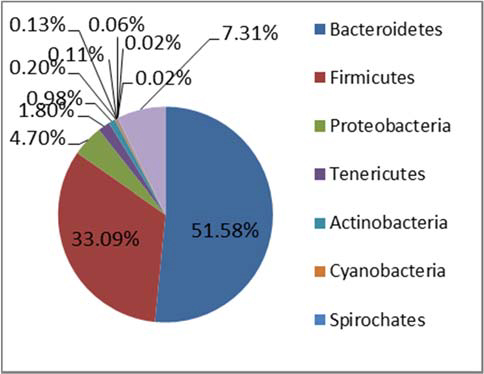
Figure 1. Phylum level taxonomic composition of the core microbiota of STEC high‐shedders and low‐shedders.
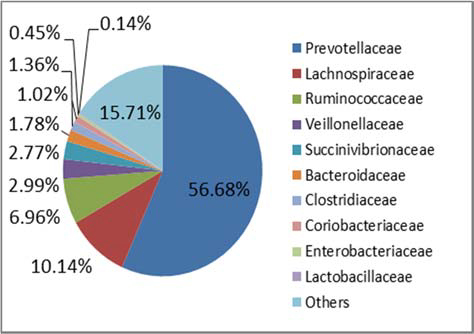
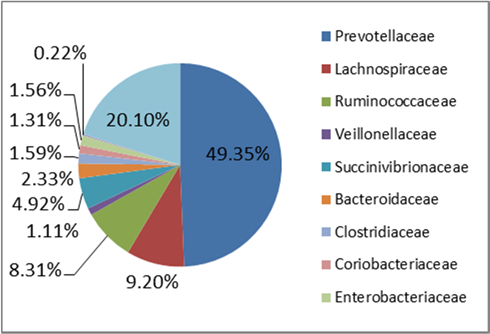
Figure 2. Family level taxonomic composition of the core microbiota of STEC high‐shedders and low‐shedders.
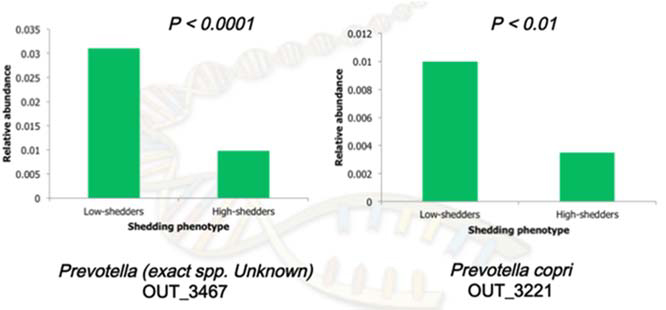
Figure 3. Differences in abundance of two of the candidate microbial species isolated as DFM.