Project Summary
Screening for Antibiotic-Resistance Genes and Class 1 Integrons in Commensal Bacteria in Agricultural and Other Environments and their Potential Transfer to Pathogenic Bacteria
- Principle Investigator(s):
- John N. Sofos
- Institution(s):
- Center for Meat Safety & Quality Food Safety Cluster, Department of Animal Sciences, Colorado State University
- Completion Date:
- 2009
Background
Antibiotic resistance in pathogenic bacteria causes difficulties in effectively treating some bacterial infections. Bacteria may acquire resistance genes through Class 1 integrons under environmental pressures. Integrons have been widely reported in clinical isolates and isolates from food animals. Recently, the presence of integrons in urban and pristine areas has drawn attention to the transfer of resistance genes between integron-associated settings. Investigating the presence of Class 1 integrons in agricultural and other environments could help us recognize resistance gene reservoirs and their movements among ecosystems in response to various pressures.
The objectives of this study were:
i. To assess the distribution of antibiotic-resistance genes and Class 1 integrons in animal fecal samples and agricultural environment samples; non-farm environment samples were tested as well, for comparison. The obtained information should provide a better understanding of the potential reservoirs for antibiotic-resistance genes present in farm and other environments.
ii. For commensal isolates that were Class 1 integron-positive, conjugation experiments were conducted in the samples in which the Class 1 integron-positive isolates were originally present to estimate their potential transfer to pathogenic bacteria under various conditions.
Methodology
A total of 330 samples were collected from cattle operations and non-farm environments including city locations and a national park in Colorado (Figure 1). The levels of tetracycline-resistant and ceftiofur-resistant bacteria populations were evaluated by both agar plating and real-time PCR methods. A total of 1,870 antibiotic-resistant isolates were purified and subjected to polymerase chain reaction (PCR) screening of Class 1 integrons. Commensal bacteria-containing Class 1 integrons were identified by 16S rRNA gene sequencing. The resistance genes within Class 1 integrons were also determined by DNA sequencing. The bacteria-containing Class 1 integrons were tested for their resistance to clinically important antibiotics. The transferability of Class 1 integrons from commensal bacteria to pathogenic bacteria was studied by conjugation experiments.
Findings
- Total and antibiotic-resistant bacteria counts determined by both culture plating and real-time PCR methods were not significantly different in soil, fecal and water samples from cattle operations and non-farm environments.
- Levels of ceftiofur resistance genes were much lower than tetracycline resistance genes.
- Class 1 integrons were detected in fecal and water samples from farm environments and city locations, and soil, feed (unused), compost and manure samples from the dairy farm, indicating that this mobile antibiotic-resistance-genetic element is widely distributed in both cattle operations and non-farm environments (Figure 2). Taxonomy of bacterial isolates containing Class 1 integrons from city locations was close to clinical bacteria which are the primary source of Class 1 integrons. Bacterial isolates containing Class 1 integrons from cattle operations had diverse taxonomic relationships.
- Class 1 integron-positive isolates from city locations displayed multi-resistance to 7-13 antibiotics tested, while Class 1 integron-positive isolates from cattle operations displayed diverse resistance patterns to 1-21 antibiotics tested.
- Most Class 1 integrons had one gene cassette belonging to the aadA family which confers resistance to streptomycin and spectinomycin.
- One dog-fecal isolate from a city dog park transferred its Class 1 integron to Escherichia coli O157:H7.
- At the molecular level of detection and characterization of Class 1 integrons, Class 1 integron-positive isolates from cattle operations displayed properties that belong to commensals naturally present in environments. Class 1 integron-positive isolates from city locations showed properties that are usually found in clinical isolates. They included isolates belonging to gamma-proteobacteria, of multiple resistances, and able for self-transferability of Class 1 integrons.
Implications
This study revealed that environmental commensal bacteria may serve as reservoirs for Class 1 integrons. Beef and dairy cattle may acquire Class 1 integron-positive isolates through their contact with the environment. Therefore, it is necessary to conduct risk assessments and explore strategies in order to minimize Class 1 integrons in cattle operations and to prevent non-transferable Class 1 integrons from becoming transferable.
The presence of Class 1 integrons in both cattle operations and non-farm environments suggests that antibiotic resistance in the environments is a complex ecological problem. Activities in cattle operations do not exert more obvious pressure than urban activities on the selection of Class 1 integrons. The antibiotic resistance phenomenon should be addressed in a way that takes into account a multitude of factors. The findings highlight the need of comprehensive studies in various environments to determine the type and level of selection pressure and its effect on the development, persistence and transmission of antibiotic resistance.
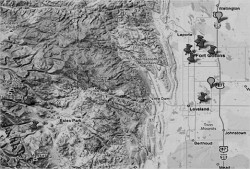
Figure 1. Sample collection sites of cattle operations and non-farm environments in Colorado.
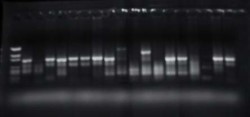
Figure 2. Class 1 integrons detected by PCR.