Project Summary
Investigation of Molecular Targets that Identify the Presence of Pathogenic Non-O157 STEC
- Principle Investigator(s):
- Joseph Bosilevac, Terrance Arthur, Norasak Kalchayanand, Tommy Wheeler, Mohammad Koohmaraie
- Institution(s):
- U.S. Meat Animal Research Center, IEH Laboratories & Consulting Group
- Completion Date:
- May 2011
Background
Non-O157 STEC are an increasing concern to regulators, beef industry and consumers. However, rapid and reliable detection of this group of organisms is lacking. STEC contain a variety of virulence factors that may serve as useful markers that allow a more narrowly focused screening program to be established. The experiments described here focus on determining which virulence genes are the best targets to meet this end.
The objectives of this study were to determine the combination of molecular markers that best indicates the presence of non-O157 STEC in a beef culture enrichment.
Methodology
PCR methods were used to identify the presence of a variety of virulence factors present in non-selective beef sample enrichments. Sponge samples were collected from hides and pre-evisceration carcasses. Only the carcass sponges were used in this analysis. Primers were identified or designed to the virulence factors of interest and standard PCR was used to determine their presence. All samples that contained Shiga toxin genes were subjected to STEC isolation by plating to washed sheep’s blood agar. All isolates were categorized as STEC (stx positive), EPEC (intimin positive) or pathogenic STEC (stx and intimin positive). The presence of each type of E. coli was correlated to the number of virulence factor genes present in the enrichments.
Findings
The virulence genes nleB, nleC, subA and espK correlated with samples that contained Shiga toxin genes (Table 1). Culture confirmation performed on Shiga toxin positive enrichments showed that as the number of virulence gene markers detected increased, the proportion of pathogenic STEC isolates recovered increased while the proportion of non-pathogenic STEC isolates recovered decreased (Figure 1).
Implications
Current STEC detection methods (FSIS method) will identify an unacceptably high number of suspect samples that will either require a lengthy confirmation (culture isolation) or be directed to thermal heat treatments at a loss of value. By using additional virulence factors as indicators for the presence of pathogenic STEC, a much smaller number of suspect samples will be identified.
Table 1. Prevalence of selected markers in enrichments identifies those that are more prevalent in STEC positive and STEC negative enrichments.
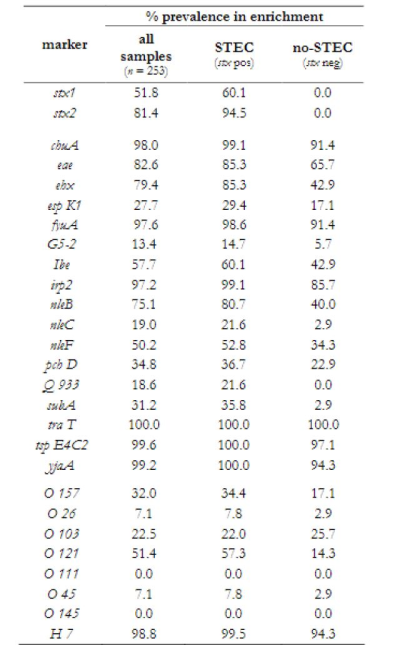
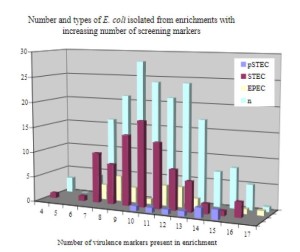
Figure 1. Enrichments that contained between 10 and 15 virulence markers yielded increasing proportions of pSTEC and decreasing proportions of non-pathogenic STEC. The prevalence of interfering EPEC was about the same throughout all the enrichments.
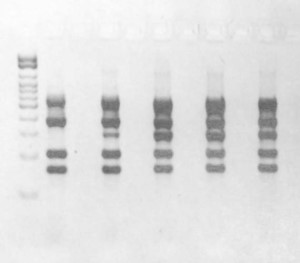
Figure 2. PCR screening results targeting Shiga toxin, intimin, nle, and subtilase genes used to identify suspect beef processing enrichments that contain pathogenic STEC.
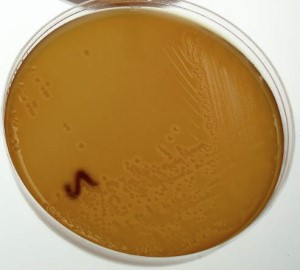
Figure 3. Characteristic growth of STEC-)121 on washed sheep’s blood agar following isolation of the STEC from a beef processing enrichment.