Project Summary
Initial Investigation of the Burden of Methicillin-Resistant Staphylococcus aureus (MRSA) in Cattle
- Principle Investigator(s):
- R. M. McCarthy1, G. H. Loneragan1, and M. M. Brashears2
- Institution(s):
- 1Department of Agricultural Sciences, West Texas A&M University
2Department of Animal and Food Sciences, Texas Tech University - Completion Date:
- 2009
Background
Antimicrobial use in animal production has been scrutinized in recent years and believed by some to be potentiating the burden of drug-resistant organisms. Some of these drug-resistant bacteria are capable of causing or complicating disease in humans. Methicillin-resistant Staphylococcus aureus (MRSA) is one such pathogen. This phenotype of S. aureus resists β- lactam drug therapy, thus leaving limited therapeutic options to treat disease caused by MRSA.
MRSA is a well-known cause of nosocomial infections worldwide, and recently, evidence has suggested it can be a community acquired and zoonotic pathogen. Consequently, public-health control of this organism poses new challenges for both human and animal practitioners. MRSA has been identified in numerous animal species including pigs, cattle, and horses.
In light of MRSA being detected in food-producing animals and reports of food borne transmission of MRSA in other countries, the researchers sought to investigate the burden of MRSA in beef cattle in the Texas High Plains in order to evaluate cattle as a potential reservoir for the pathogen and subsequently a possible food-borne source of MRSA and assess the susceptibility of methicillin-susceptible S. aureus (MSSA) recovered from cattle of the Texas High Plains.
Methodology
Three samples (nasal, hide-dorsal midline, and hide-perianal) were collected from 301 cattle (111 feedlot, 81 cull dairy, 91 cull beef cows, and 18 cull beef bulls) of various sources using sponges pre-hydrated with tryptic soy broth supplemented with 7% NaCl. Broth was incubated at 35°C for 20 hrs. Each sample was then streaked using a 3-step dilution method onto two MRSA selective agar plates and incubated at 35°C for 24 hrs and two coagulase-mannitol agar plates and incubated at 35°C for 18 hrs. Suspect colonies were streaked on to tryptic soy agar for further evaluation using catalase and coagulase testing. Catalase and coagulase-positive isolates were subjected to automated biochemical confirmation, and antimicrobial susceptibility was determined using micro-broth dilution in addition to disk diffusion techniques.
Findings
Samples were collected from 111 beef-type cattle housed in feedlots. Of these, 60 were close to harvest and the remainder was new arrivals. Eighty-one samples were collected from dairy-type cattle at a regional livestock auction facility; these animals were culled from their herd of origin for poor productivity reasons. One hundred and nine samples were collected from beef-type animals at the same sale barn.
MRSA was not recovered from any of the samples collected (0.0%, n = 0; 95% CL = 0, 1.3%). Of the 301 animals sampled, MSSA was isolated from 14.3% (n = 42) of cattle resulting in 82 isolates. MSSA was recovered from 21.1% of beef-type and 2.5% of dairy-type cattle from the auction, and 16.2% of feedlot-origin cattle. Overall, samples collected from the nares yielded the greatest recovery of S. aureus (58.7%).
Susceptibility results for disk diffusion are still being analyzed. Sensititre results are reported here. Of the isolates, 17.1% were susceptible to all drugs tested. The most commonly observed resistance was to chloramphenicol (53.7%) followed by daptomycin (50%), tetracycline (1.2%) and erythromycin (1.2%). No isolate was resistant to four or more drugs. The phenotypes observed are presented in Figure 1.
Implications
While S. aureus is not uncommon on the surface of animals, the burden of MRSA carried by cattle in the Texas High Plains appears to be negligible, and considerable increases in sample size are needed to detect such a rare event. Given that the most productive site of isolate recovery was nasal swabs, future studies designed to characterize S. aureus isolates may be best focused on this site with increases in number of animals sampled. All MSSA isolates recovered in the study were broadly susceptible to antimicrobial drugs. The data reported herein provide evidence that S. aureus found on surfaces of cattle in the Texas High Plains are of little, if any, food-borne or direct-contact consequence.
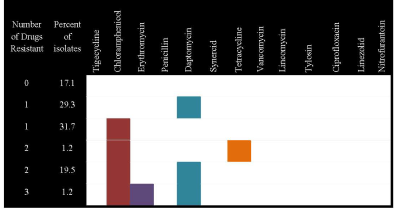
Figure 1. Antimicrobial resistance phenotype, percentage of isolates, and number of drugs to which they were resistant.