Project Summary
Hide to Carcass Transfer of E. coli O157:H7 and Salmonella in Small-scale (less than 1000 head/day) Beef Processing Plants
- Principle Investigator(s):
- J. M. Bosilevac, T. M. Arthur, M. N. Guerini, D. M. Harhay, N. Kalchayanand, D. A. King, S. D. Shackelford, T. L. Wheeler and M. Koohmaraie
- Institution(s):
- U.S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2008
Background
There is ample data on the prevalence and levels of E. coli O157:H7 and Salmonella found during the different steps of slaughter at large US beef processing plants. However, a significant portion of the U.S. beef supply passes through small-scale processing plants, for which little data is available. For this study “small-scale processing plants” is defined as processors who slaughter less than 1,000 cattle a day. The small plants typically do not have the resources available to the large processors. Therefore, interventions and technology to reduce hide-to-carcass transfer of pathogens may be limited. Small processors are eager to provide the safest beef possible, and in need of evaluation to determine the best processes to put into place. The results of this work will directly help the small processors involved evaluate their practices, while the combined data will answer questions regarding the levels of E. coli O157:H7 and Salmonella encountered by small scale U.S. beef processors.
The stated objectives for this work were to:
- Determine the prevalence and enumerate E. coli O157:H7 on hides and pre-evisceration carcasses of cattle slaughtered at small processing plants.
- Determine the prevalence and enumerate Salmonella on hides and pre-evisceration carcasses of cattle slaughtered at small processing plants.
Methodology
Design. Seven small-scale processors from across the country were visited and samples were collected. Ninety-five (95) hide samples (1,000 cm2) and 95 pre-evisceration carcass samples (8,000 cm2) were collected each day for 3 days at each processing plant. An additional 5 samples/day were collected for use as controls. Samples were collected and shipped to MSQRU for analysis.
Enumeration. Samples were enumerated for E. coli O157:H7 and Salmonella by established USMARC methods. Briefly, the methods consisted of spiral dilution plating hide samples directly onto selective media, ChromAgar O157 and XLD, for E. coli and Salmonella respectively. Carcass samples were enumerated using hydrophobic grid membrane filters placed on the same media.
Culture isolation and confirmation of E. coli O157:H7. The prevalence E. coli O157:H7 was determined by established USMARC methods that employ a nonspecific enrichment in tryptic soy broth (TSB) followed by immunomagnetic separation (IMS) of E. coli O157 and plating to ChromAgar O157. Suspect colonies were screened using Oxoid DrySpots for O157, then confirmed to be E. coli O157:H7 that contained at least one virulence factor (stx1, stx2, or eae) by multiplex PCR.
Culture isolation and confirmation of Salmonella. The prevalence of Salmonella was determined by established USMARC methods. Salmonella isolation is run concurrently to E. coli O157:H7 detection and uses the same nonspecific enrichment in TSB. The Salmonella is concentrated by IMS, and the IMS beads are selectively enriched for Salmonella by incubation in Rappaport-Vassiliadis-Soya (RVS) broth before plating to Hektoen Enteric agar and Brilliant Green medium with Sulfadiazine. Suspect colonies were isolated and confirmed to be Salmonella by PCR for the invA gene.
Data analysis. The prevalence and levels of E. coli O157:H7 and Salmonella on hides and carcasses were used to determine the rate of hide-to-carcass transfer of the pathogens at each small processor. For this and all subsequent reports, the values for each plant have been combined and averaged for a composite example of small U.S. beef processors.
Findings
Over the course of this project, samples were collected for 3 days at 7 different small processing plants. The samples were enumerated for E. coli O157:H7 and Salmonella as well as overall prevalence on hides and carcasses. The results shown in Table 1 provide the number and percentage of samples of each type that were enumeration positive, (that is; samples with 40 or more CFU/100 cm2 on hides or 0.4 CFU/100 cm2 on pre-evisceration carcasses) and prevalence positive as determined by culture isolation. On a day-by-day basis, hide prevalence of E. coli O157:H7 ranged from 28% to 100% positive, while Salmonella prevalence on hides ranged from 57% to 100%. The range of E. coli O157:H7 prevalence on carcasses was even more marked, ranging from 0% to 93% on different days at different plants. Salmonella had a similarly wide day-to-day prevalence ranging from 16% to 99%.
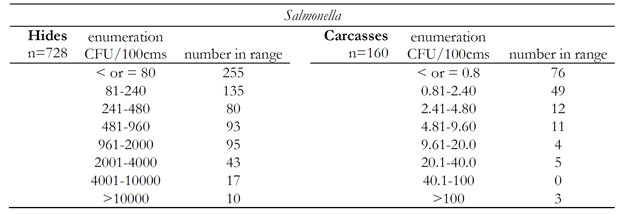
The enumeration results for E. coli O157:H7 are shown in Table 2. Generally, more than half of the samples that were enumeration positive were positive at a low level, which was just above the detection limit of the enumeration methods. This observation held for enumeration of Salmonella as well (Table 3). It was noted that higher enumerable levels of pathogen on hides positively correlated to pathogen on corresponding carcasses. Therefore, reducing hide levels of pathogens should be a priority for small processors.
Implications
Data showed higher levels of pathogens on hides positively correlated to higher levels of pathogen on carcasses. Therefore, reducing hide levels of pathogens should be a priority for small processors. Additionally, through the sharing of results and observations, each small processor was able to rapidly implement changes to their practices to improve beef safety. Examples of such changes included improved hide removal steps, carcass steam vacuuming and trimming, and final trim sampling for test-and-hold.
Table 1. Number and percent positive of samples for E. coli O157:H7 and Salmonella on hides and pre-evisceration carcasses by enumeration and culture isolation (prevalence positive).
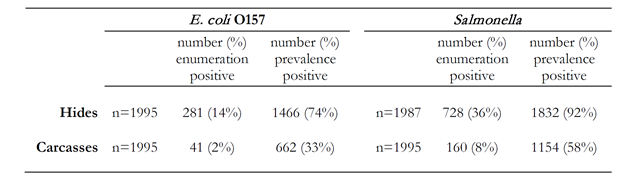
Table 2. Number of E. coli O157:H7 enumeration positive samples in each range shown for hides and carcasses.
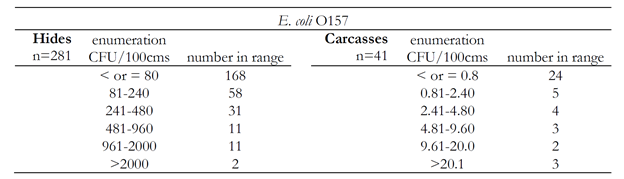
Table 3. Number of Salmonella enumeration positive samples in each range shown for hides and carcasses.