Project Summary
Detection of Pathogenic and Shiga Toxin-Producing Escherichia coli from Beef by Multiplex PCR
- Principle Investigator(s):
- Wendy Maduff, Gina Bellinger, and Wendy Warren
- Institution(s):
- Food Safety Net Services
- Completion Date:
- 2009
Background
Non-O157 Shiga toxin-producing Escherichia coli (non-O157 STEC) have been identified as an emerging public health issue. The six most prevalent serotypes, including O26, O45, O103, O111, O121, and O145, accounted for 71% of the 940 isolates associated with illness from 1983 to 2002. Some non-O157 STEC possess the same range of virulence factors as E. coli O157:H7 capable of causing serious illnesses, or death. For the above reasons and its high prevalence levels in other countries, STEC is on the emerging pathogen watch list. It is difficult to distinguish pathogenic non-O157 STEC strains from non-pathogenic E. coli because the former rarely possess any distinguishing phenotypic or biochemical characteristics from the latter. The lack of reliable and validated laboratory methods for testing various food matrices has meant that food is not routinely tested for non-O157 STEC. If non-O157 STEC is identified as a potential serious threat due to high numbers in cattle, the industry will require a reliable tool for detection and control.
The objective of this study was to develop a real-time multiplex PCR test that reliably identified the six most prevalent Shiga toxin-producing E. coli in the United States from beef.
Methodology
Fresh cultures of each E. coli strain (Table 1) were prepared for DNA isolation and optimization of assay conditions or were directly inoculated into fresh beef trim and ground beef for detection.
Beef samples, purchased from a local grocery store, were inoculated one strain at a time with 100, 101, 102, or 103 colony forming units per sample, and were incubated in TSB at a 1:10 ratio at 37°C for 18 hours.
A total of nine genetic targets were ultimately selected for PCR-based detection, including one for each of the top six STEC (O26, O45, O103, O111, O121, and O145), O157:H7, eae (relevant virulence gene), and gapA (E. coli associated gene for control). The real-time PCR was optimized for each target using purified DNA, then tested lysed on pure culture, and then detected from inoculated raw beef trim and ground beef.
Findings
Genetic targets were successfully detected from purified DNA, lysed cells from pure cultures, and for a target in beef trim and for some targets in ground beef protease treated 18-hour enrichments. Images shown in Figures 1 and 2 are representative of the assays ability to detect high levels of DNA efficiently. These images show serial dilutions of purified DNA; the assay makes a ‘positive’ identification when the line crossed the horizontal green or red arrow.
The assay was tested on protease treated enrichment, which proved to be the most challenging. The enrichment from the beef trim proved to be an easier to test than the enrichment from ground beef, likely due to higher fat in the ground beef. Both uninoculated matrices were positive for GAPDH, which is not surprising as gapA is found in most E. coli, which is frequently in beef. The eae assay was only positive from the ground beef enrichment; the Ct value was 60, therefore, either there was a very low level of eae in the enrichment (not necessarily from E. coli) or it was a false-positive. Further optimization of the assay is warranted to improve the efficiency of detection from raw beef matrices.
Implications
This assay may prove to be very important if the government mandates testing for these strains as well as a potentially valuable processing tool, as STEC might be a good indicator for the presence of E. coli O157:H7.
Table 1. E. coli strains used for assay design.
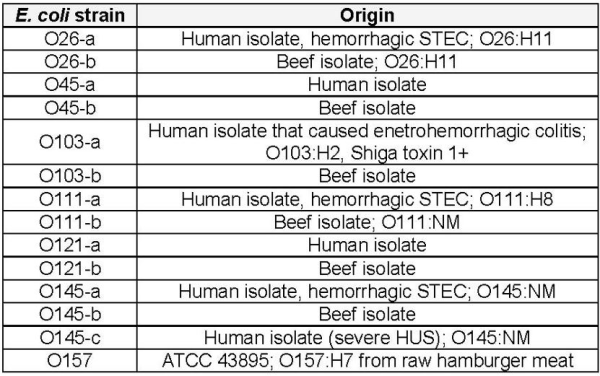
Figure 1. The O121 assay results from serial dilutions of purified O121 DNA.
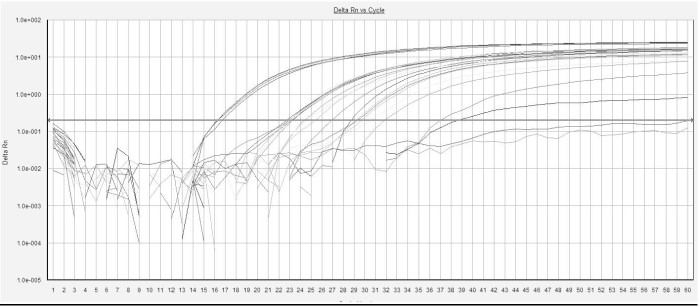
Figure 2. The O145 assay results from serial dilutions of purified O145 DNA.