Project Summary
Colonization Characteristics of Bovine Recto-anal Junction Tissues by Escherichia coli O157:H7
- Principle Investigator(s):
- James L Bono, Terrance M. Arthur, and Tommy L. Wheeler
- Institution(s):
- U.S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2008
Background
Escherichia coli O157:H7 is currently a major food safety concern for the meat industry. While post-harvest interventions such as hide-wash cabinets and test-and-hold have decreased the prevalence in meat products, pre-harvest interventions to reduce the prevalence in feces and on hides have not been as successful. One possible pre-harvest intervention approach would be to target the recto-anal junction (RAJ), a colonization site of E. coli O157:H7 in the bovine gastrointestinal (GI) tract. A better understanding of colonization at the RAJ could lead to interventions that block or reduce adherence of bacteria at this site.
Recently, a site of E. coli O157:H7 colonization was identified in the bovine intestinal tract. This site was located at the RAJ. Prior to this finding, the development of interventions targeted at reducing E. coli O157:H7 in the host animal was confined to a “trial-and-error” approach. In this methodology, potential intervention strategies were chosen based on speculation of efficacy and they were applied to the animal to determine if indeed they were effective. With the findings of the RAJ colonization site, there can now be more rigorous selections and screenings of potential interventions, thus, insuring greater success of the interventions. Probiotic development is a prime example. Traditionally, organisms were chosen based on their ability to inhibit the growth of E. coli O157:H7. However, many of these organisms were not effective in vivo due to changes in pH, density, and indigenous microflora. With the development of specific binding and competition assays, it will be possible to select for those organisms that directly compete with E. coli O157:H7 for colonization.
The locus of enterocyte effacement (LEE) is an E. coli O157:H7 pathogenicity island. tir and eae are two LEE-encoded genes responsible for the tight bacterial association with the intestinal mucosa in humans and cattle. The eae-encoded ligand protein intimin is located on the bacterial outer membrane; whereas, the intimin receptor protein tir is translocated into the epithelial cell by type III secretion and integrated in the host cell membrane. Isolates with deletions in the tir or eae gene have reduced binding to epithelial cells in culture, reduced virulence in animal models, and reduced length of fecal shedding in cattle.
The researchers recently identified a single nucleotide polymorphism (SNP) in the tir gene that associates with the potential of an organism to cause disease in humans. Ninety nine percent of human E. coli O157:H7 isolates have a T nucleotide at this site (tir sp-1 T allele), while only 1% of the human isolates have an A nucleotide at this site (tir sp-1 A allele). In cattle, 56% of the E. coli O157:H7 isolates have the tir sp-1 T allele, whereas, 44% have the tir sp-1 A allele. Also, in vitro cell adherence to human (Hep-2) or bovine (MDBK) epithelial cells associates with the tir sp-1 alleles. Isolates with the tir sp-1 T allele adhere in cell culture significantly more than isolates with the tir sp-1 A allele (Figure 1). It was hypothesized that isolates with the tir sp-1 T allele will colonize the RAJ and result in a longer duration of fecal shedding compared to isolates with the tir sp-1 A allele.
The stated objectives for this work were to:
A. Develop an in vitro model system for the binding of E. coli O157:H7 to bovine recto-anal junction (RAJ) tissue.
B. Characterize the binding of E. coli O157:H7 to the bovine RAJ.
C. Characterize the shedding patterns of E. coli O157:H7 with different tir alleles in cattle colonized at the RAJ.
Methodology
A. In vitro model system.
Collection of bovine intestinal epithelial cells. Sections (0.5 m) of bovine colon, up to and including the anal canal, were collected immediately following evisceration at a local, commercial beef processing plant. The colon sections were transported back to the lab in insulated carriers to prevent rapid cooling. At the lab, the colons were opened and the RAJ identified. Sections of the RAJ and upper colon epithelia were removed. The tissue sections were processed according to a previously published protocol. Briefly, the tissues were washed three times in minimal essential medium (MEM)-HEPES. The RAJ and upper colon tissue section were scraped with a plastic spoon to dislodge the epithelial cells. Tissue scrapings were re-suspended in 2 mL of MEM-HEPES and stored at 4°C until use.
Bacterial strains. Bacterial strains (two Shiga toxin-containing E. coli O157:H7 and two generic E. coli strains of the O157 serogroup) were grown 16-18 h at 37ºC with shaking in MEM-HEPES.
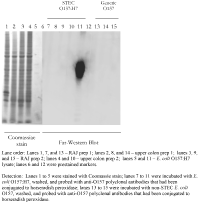
Far-Western blotting. Bovine intestinal epithelial cells were sonicated 3 × 30 s each using a microtip sonicator. Cell lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE). The proteins were electrophoretically transferred onto Polyvinylidene fluoride transfer membrane. The membrane was blocked by incubating in MEM-HEPES with 2% nonfat dry milk (NFDM) for 1 h at room temperature. Nonadherent bacteria were removed by washing them three times in phosphate buffered saline (PBS), and adherent bacteria were detected with an anti-O157 horseradish peroxidase-labeled polyclonal antibody and chemiluminescent substrate.
In vitro intestinal epithelial binding assay. Intestinal epithelial cells (50 μl) were mixed with 150 μl of MEM-HEPES in wells of NUNC Maxisorp microtiter plates. The cells were loaded as follows: four columns of eight wells contained colon cells, while another four columns of eight rows contained RAJ cells. The plates were washed five times with PBS. The blocking solution was removed, and the wells were washed five times with PBS. Bacteria (100 μl) were added to each well and incubated for 1 h with shaking at 37°C.
Nonadherent bacteria were removed by washing them five times in PBS, and adherent bacteria were detected with an anti-O157 horseradish peroxidase-labeled polyclonal antibody and chemiluminescent substrate.
B. Procedures for testing E. coli O157:H7 tir alleles for colonizing the bovine RAJ mucosa. The RAJ colonization experiment was performed in collaboration with the University of Idaho which had the facilities to perform the animal challenge study.
Animals. To assure rumen maturity, five-month-old steers were used in all experiments. Animals were housed in groups of four and inoculated with either E. coli O157:H7 isolates that contain the tir sp-1 A or T allele by rectal application (see below). Four animals were used to screen two isolates from each allelic group (n = 16 animals).
Bovine rectal administration of E. coli O157:H7 isolates. Animals were housed in quarantined facilities in which feed, water, bedding, manure, and other waste would not be in contact with any non-experimental animals. All personnel followed strict biosafety procedures and all procedures were approved by the Institutional Animal Care and Use and Biosafety Committees. Animals were inoculated by rectal application of E. coli O157:H7 tir sp-1 A or T allelic isolates. Briefly, feces was manually removed from the terminal rectum and a 10 cm by 3.5 cm diameter cylindrical sponge with a wooden handle was saturated with 10 mL of the E. coli O157:H7 culture containing 1010 CFU, inserted into the terminal rectum, and rubbed against the RAJ mucosa. After swabbing, defecation was prevented for ~10 min by holding the steer’s tail firmly against the anus. This ensured that the inocula would be retained in the rectal lumen for a sufficient period of time for bacteria to associate with the mucosa.
RAMS culture technique for E. coli O157. To determine the ability of wild-type E. coli O157:H7 to colonize cattle, a recently developed RAMS culture technique that is superior to fecal culture and immunomagnetic bead separation techniques was used. Rather than culture E. coli O157:H7 from fecal material that contains numerous competing bacteria, the recto-anal junction mucosa was swabbed and cultured. Briefly, RAMS samples were obtained by inserting a sterile foam-tipped applicator approximately 2 to 5 cm into the anus and, using a rapid in and out motion, the entire mucosal surface of the RAJ was swabbed. Each RAMS sample was placed into a culture tube containing 3 mL trypticase soy broth (TSB) and kept on ice until processed in the laboratory within 2 h of collection.
Findings
An in vitro binding assay that is rapid and specifically mimics the adherence of E. coli O157:H7 to the site of bovine colonization at the RAJ would be a major step in elucidating the mechanisms involved in E. coli O157:H7 colonization and shedding cycles in cattle. Two approaches were taken to investigate the adherence of E. coli O157:H7 to RAJ cells. In the first approach, Far-Western blotting was used in an attempt to identify protein-protein interactions that mediated binding (Figure 1). Colon and RAJ cell lysates were loaded onto polyacrylamide gels and the individual proteins were separated by electrophoresis. An E. coli O157 lysate was also loaded as a detection control. The cellular proteins were denatured during the electrophoresis process and then renatured during the transfer and blocking steps. The renatured proteins were probed with viable cells of either STEC O157:H7 or generic O157. When probed with the anti-O157 antibody, no differences in binding could be detected with this system between STEC O157:H7 and generic O157.
In the second approach, primary cell cultures of bovine intestinal epithelial cells, colon, and RAJ were bound to microtiter plate wells and incubated with either STEC O157:H7 or generic O157 strains (Figure 2). In this assay, high levels of binding were detected, but the binding was not specific for E. coli O157:H7 to RAJ cells. The STEC O157:H7 cells bound equivalently to colon and RAJ cells. In addition, generic O157 cells had the same level of adherence as did the STEC O157:H7 cells.
Four groups, two groups with the tir T allele and tir A allele, each with four, three-month-old steers were challenged with one of four strains of E. coli O157:H7. RAJ mucosa swabs were cultured twice a week for E. coli O157 and means were calculated for each group of steers to determine shedding enumeration and duration. All isolates recovered from the experiment were confirmed as the isolate used to inoculate the challenge animal by PCR for the O157 O-antigen, tir allele genotyping and PFGE. There was no statistical difference between the shedding enumeration and duration of the isolates used in this study (Figure 3).
The premise of this work was the difference in adherence of the two different tir allele isolates to bovine MDBK epithelial cells in an in vitro assay. The tir allele most commonly found in isolates from humans adhered to the MDBK cell in the in vitro assay 3.5 times more than the other allele. However, this increased adherence was not observed in an in vivo shedding model. There are several reasons why the in vitro assay did not correlate with the in vivo model. Most notably, the growing environments were different. The in vitro model lacks the microbial background that is found in the lower bovine GI tract. Bacteria to bacteria signaling and competition for nutrients can alter the expression of bacterial genes. This could allow for E. coli O157:H7 to adhere to the RAJ regardless of the tir genotype. Evidently, factors in the in vitro cell culture assay allow expression of genes that associate the number of adherent bacteria with the potential for the isolate to cause disease in humans.
Implications
E. coli O157:H7 resides in the intestinal tracts of cattle. Knowledge of the manner in which these bacteria attach to the cattle intestinal cells is crucial to the development of methods for eliminating E. coli O157:H7 in the cattle population. Experiments were carried out to interrogate the binding of E. coli O157:H7 to the intestinal tract cells of cattle did not detect differences between generic and pathogenic O157. In an animal model for recto-anal junction colonization, there was no difference in shedding duration or enumeration between E. coli O157:H7 isolates that are associated with human disease and isolates that are not.
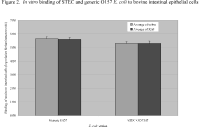
Figure 2. In vitro binding of STEC and generic O157 E. coli to bovine intestinal epithelial cells.
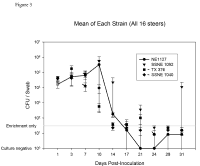
Figure 3. Mean CFU/Swab for each E. coli O157:H7 strain. Four groups, each with four, 3-month-old Holstein steers were challenged with one of four strains of E. coli O157:H7, RAJ mucosa swabs were cultured twice a week for E. coli O157 and means were calculated for each group of steers. Error bars indicate ± S.E. E+, positive by enrichment only.