Project Summary
Effects of Antimicrobial Interventions on Multi Drug-Resistant (MDR) Salmonella
- Principle Investigator(s):
- Terrance Arthur, Joseph Bosilevac, Dayna Brichta, Michael Guerini, Norasak Kalchayanand, Steven D. Shackelford, and Tommy Wheeler
- Institution(s):
- U. S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2007
Background
Salmonellosis is the second most common foodborne illness in the United States. An estimated 1.4 million cases occur annually in the United States; of these, approximately 30,000 are culture-confirmed cases reported to the Centers for Disease Control and Prevention, resulting in approximately 582 deaths. Accordingly, Salmonella is the leading cause of foodborne infections with lethal outcomes in the United States. The most common vehicles of transmission are meat and poultry products, eggs, and egg products that contain Salmonella. Two thousand serotypes of Salmonella are believed to cause human disease, but the majority of disease cases are caused by just five serotypes.
Salmonella enterica serovars Typhimurium and Newport are commonly identified in bovine clinical samples and in 2003 represented 11% and 30% of the reported bovine isolates respectively. S. Typhimurium and S. Newport are also two of the top three disease-causing Salmonella serovars in the United States and were responsible for 20% and 11%, respectively, of the reported cases of salmonellosis in 2003. The largest age group to be affected by salmonellosis consists of children under 5 years of age, who represent approximately 25% of the reported cases each year. Salmonellosis in children is often invasive and requires antibiotic treatment for recovery. As such, it is concerning that in recent years, there has been a marked increase in the number of multi drug-resistant Salmonella isolated in clinical settings. S. Typhimurium with the ACSSuT resistance pattern (ampicillin, chloramphenicol, streptomycin, sulfisoxazole, and tetracycline), and S. Newport with the MDR-AmpC resistance pattern (Multi Drug-Resistant S. Newport that are resistant to at least 9 of 17 antibiotics tested including: amoxicillin/clavulonic acid, ampicillin, cephalothin, ceftiofur, cefoxitin, ceftriaxone, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline), were two of the most common multi-drug resistant Salmonella phenotypes isolated in 2002. In 2002, S. Newport was the reported cause for 4204 (13%) of the 32,308 cases of salmonellosis. Approximately 6% of the S. Newport clinical isolates were sent to the CDC for antibiotic susceptibility testing and 22% (53 of 239) were found to be multi drug-resistant. The increasing number of S. Newport infections appears to be associated with the emergence and rapid dissemination of the multi drug-resistant strains of MDR-AmpC Newport.
As the selective pressure exerted by antimicrobial drug use in humans and agriculture persists, the continued and increasing prevalence of multi drug resistance among Salmonella is likely to be observed. It is not known if strains that are resistant to multiple antibiotics possess traits that also would allow them to survive antimicrobial interventions found in beef processing plants.
The stated objectives for this work were:
- Determine efficacy of current beef processing antimicrobial interventions in reducing MDR Salmonella levels on beef carcasses.
- Determine efficacy of current beef processing antimicrobial interventions in reducing non-MDR Salmonella levels on beef carcasses.
- Compare effects of antimicrobial interventions on MDR Salmonella with the effects on non-MDR Salmonella. Both human disease-associated and non-human disease-associated, were added to the experiment.
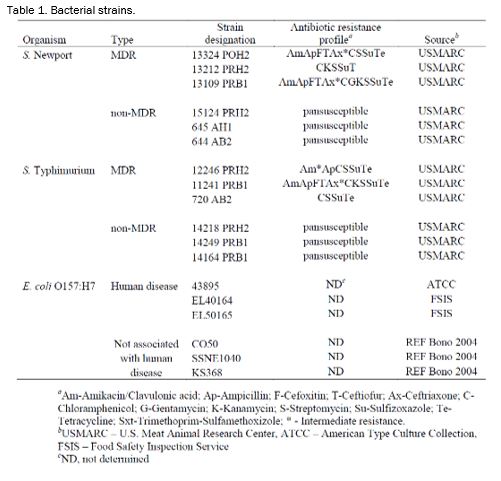
Bacterial strains. The strains used in this experiment are listed and described in Table 1. Salmonella Newport and Salmonella Typhimurium were each separated into MDR and non-MDR types. E. coli O157:H7 strains were subdivided into human disease associated (HDA) and non-HDA types (Table 1). Salmonella Newport and Typhimurium and E. coli O157:H7 strains (Table 1) were grown individually overnight in TSB. The following morning, the strains were mixed into cocktails by organism type. After mixing, the strain cocktails were diluted in BPW to ~ 1 × 108 CFU/mL. The diluted cocktails were stored on ice during the experiment. S. Typhimurium strains were not included in the acetic acid treatments. E. coli O157:H7 strains were not included in the ozone treatments.
Flank tissue. Pieces of flank tissue (~ 700 cm2) were collected from beef carcasses at a local processing plant. The tissues were obtained within 15 min post-exsanguination prior to entry of the carcasses into the antimicrobial wash cabinet. The tissue sections were transported back to the U. S. Meat Animal Research Center in insulated carriers to prevent rapid cooling. Four 100-cm2 areas were marked on each flank section with edible ink using a sterile, cotton-tipped swab and a stainless steel, 100-cm2 template before inoculation. Each 100-cm2 area was divided into four 25-cm2 areas. Each 100-cm2 section was inoculated with 500 ul of a strain cocktail at ~ 1 × 108 CFU/mL for a final concentration of 5 × 105 CFU/cm2. Three flanks were used per organism for each treatment giving twelve replications per organism per treatment.
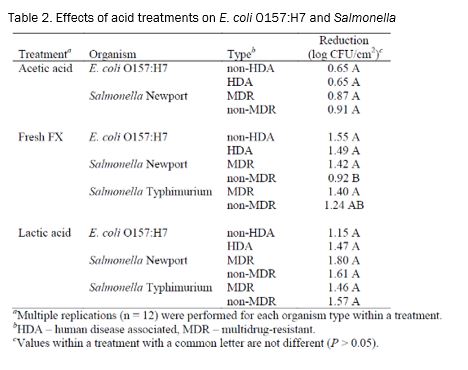
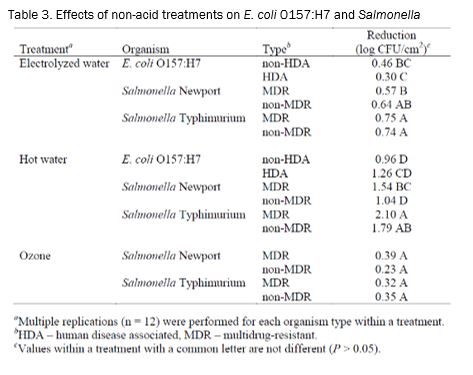
Treatment. Following inoculation, the flank sections were incubated for 15 min at room temperature to allow for bacterial attachment. Then a wash with water (25±2oC) at 45 psi was applied for 10 sec to remove any unattached bacteria. Excess water was allowed to drip off for 30 sec. After the water wash, the various treatments were applied (Table 2) for 20 sec. The treatments consisted of acetic acid (2%; Sigma, St. Louis, MO), electrolyzed oxidizing (EO) water (acidic; Electric Aquagenics, Kennesaw, GA), FreshFX® (1:50; SteriFX, Inc., Shreveport, LA), hot water (74°C at the nozzles), lactic acid (2%; Sigma), and ozonated water (6.0 ppm; Ozone International, Bainbridge Island, WA). Excess liquid was allowed to drip off for 30 sec following each treatment. Spray washes were conducted in an insertable pod in a laminar air flow hood (Cutter’00). The washer had a spray nozzle oscillation speed of 60 cycles/min.
Sampling: After each step of the process (inoculation and attachment, water wash, and treatment) 25-cm2 sections of surface tissue was excised. From each 100-cm2 section 25-cm2 pieces were excised: 1. control: removed following inoculation and attachment, 2. wash: removed following the water wash, and 3. treatment: removed following the treatment. Each 25-cm2 section was placed into a sterile Whirl-Pak bag.
Sample processing. Each bag received 25 mL of TSB-PO4 (tryptic soy broth supplemented with 0.017M KH2PO4 and 0.072M K2HPO4, pH 7.2±0.1) and was homogenized for 1 min at 9 strokes/sec using a stomacher. After stomaching, the sample contents were serially diluted 10-fold and the appropriate dilutions were plated onto XLD agar and ntChromagar (CHROMAgar O157 supplemented with 5 mg/L novobiocin and 1.0 mg/L of potassium tellurite) plates using a Spiral Plater. Plates were 4 incubated for 18-20 h at 37°C. All colonies were manually counted. Colony counts were log transformed for analysis.
Statistical analysis. Log reduction data were analyzed by analysis of variance for a completely randomized design. Main effects were strain within organism combinations. Least squares means were separated using the PDIFF option (a paired t-test) when the ANOVA was statistically significant. A predetermined probability of Type I error (α) of 0.05 was used for all determinations of statistical significance.
Findings
Acid (acetic acid, lactic acid and Fresh FX®) and non-Acid (electrolyzed water, hot water, and ozone) antimicrobial interventions were used to evaluate the reductions of E. coli O157 and Salmonella. The E. coli O157:H7 strains were subdivided into those strains having a genetic polymorphism frequently associated with strains causing human disease and those strains that do not have the particular polymorphism and are not typically found to cause human disease (Table 1). The Salmonella strains consisted of both S. Typhimurium and S. Newport serovars with each serovar being further subdivided into multidrug-resistant and susceptible categories.
The experiment described herein was designed to allow a comparison of the survival of the various strain types when a particular antimicrobial intervention was applied. The design of this experiment does not allow for the comparison of the efficacy of the various intervention types among themselves. Hence, these data should not be used as selection criteria for intervention methods.
Of the three acid treatments tested, there were no differences (P > 0.05) detected between the ability of various organisms or types to survive when exposed to acetic and lactic acid treatments (Table 2). There were differences in survival when the strains were treated with Fresh FX as non-MDR Salmonella was reduced less than any other organism x type combination.
When the non-acid treatments were evaluated, no differences (P > 0.05) in reductions were detected for Salmonella following treatment with ozone (Table 3). The electrolyzed water and hot water treatments both resulted in significant differences in strain survival. Following the electrolyzed water treatment, the Salmonella strains incurred greater reductions than did the E. coli O157:H7 HDA strains. MDR and non-MDR S. Typhimurium were reduced slightly more than the S. Newport MDR strains. Hot water produced the largest magnitude difference between strains of all of the treatments. When treated with hot water, the reduction of MDR S. Typhimurium was over 1 log greater than the reduction on non-HDA E. coli O157:H7. In addition, the reduction due to hot water of S. Newport MDR was greater than S. Newport non-MDR.
Multi drug-resistant Salmonella are viewed as emerging pathogens. These organisms are constantly acquiring new genetic sequences from other bacteria as well as their local environment. These genetic acquisitions have conferred abilities on some strains to be resistant to various antibiotics. It has been determined that the transferable DNA coding for resistance to one antibiotic frequently codes for resistance to several other antibiotics. Information has been lacking in regard to whether strains possessing multi-drug resistance properties also have the ability to resist the killing effects of the antimicrobial interventions used in beef processing.
Implications
The data presented here show that MDR Salmonella are reduced as effectively as E. coli O157:H7 when treated with antimicrobial interventions currently in use at most U.S. beef processing plants. Through this research project it has been determined that the antimicrobial interventions currently in place in beef processing facilities are adequate for reducing the foodborne pathogen loads of beef carcasses contaminated with MDR Salmonella.