Project Summary
Antibiotic and Disinfectant Susceptibility Profiles of Escherichia coli O157:H7 in Cattle Feces, Hide, Carcass, and Ground Meat Isolates from the United States
- Principle Investigator(s):
- Ross Beier1, T. Poole1, Dayna Harhay2, and Robin Anderson1
- Institution(s):
- 1USDA ARS, Southern Plains Agricultural Research Center
2USDA, ARS, U.S. Meat Animal Research Center (USMARC) - Completion Date:
- 2010
Background
Escherichia coli can cause serious illnesses or fatalities in the elderly and immunocompromised humans. The Centers for Disease Control and Prevention (CDC) estimated that each year in the United States over 62,000 human illnesses, over 1,800 hospitalizations, and 52 deaths are caused by foodborne transmission of E. coli O157:H7. The contamination of meat products with E. coli O157:H7 resulted in recall of over 1 million pounds of meat in 2005. Over 61.6 million pounds of meat products were recalled in the decade prior to 2005 because of contamination from E. coli O157:H7. Strategies used to control bacteria from the farm-to-table include the use of biocides in the form of antiseptics and disinfectants. Biocides are routinely used in animal production, the food processing industry, veterinary medicine, human medicine, consumer’s homes, and are composed of a variety of active ingredients in multiple combinations.
There is very limited information on disinfectant susceptibility of pathogens isolated from food-producing animals, and whether the evaluated bacteria have mechanisms that link resistance to antibiotics with the resistance to disinfectants. For instance, it was previously shown that beta-hemolytic E. coli isolated from neonatal swine with diarrhea had chlorhexidine resistance that was linked to resistance to the antibiotics gentamicin and streptomycin. Also, a large percentage of vancomycin-resistant Enterococcus faecium (VRE) isolated from community wastewater in Texas were found to have triclosan resistance higher than the levels of triclosan commonly found in household dish soap and in septic bath wash used in the hospital setting. Therefore, the disinfectant resistances found in pathogenic bacteria may allow them to persist in places treated with disinfectants that were assumed to be free of pathogens.
During this study a total of 344 E. coli O157:H7 isolates from feedlot cattle feces (97 isolates from Kansas, Oklahoma, Nebraska, and Texas), hides and carcasses (47 isolates from Kansas, Montana, Texas, and Utah), 70 isolates from hides and 30 isolates from previseration samples from culled cows (from 4 different regions), and 100 ground meat isolates obtained from processing plants were defined by disinfectant and antibiotic susceptibility testing, and further by determining if certain genes were present associated with disinfectant resistance and with the expression of larger regions of DNA often associated with antibiotic resistance.
Methodology
All isolates were confirmed as E. coli O157:H7 using R&F Escherichia coli O157:H7 Chromogenic Plating Medium (M-0300) obtained from R&F Products. Minimum inhibitory concentrations (MICs) were determined by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) standards for antimicrobial susceptibility testing. Antibiotic resistance was determined using the Gram-negative plates CMV1AGNF and CMV1DW obtained from Trek Diagnostic Systems Inc. The antibiotics tested were amikacin, ampicillin, amoxicillin/clavulanic acid, cefoxitin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim/sulfamethoxazole, and the fluoroquinolones on the CMV1DW plate were ciprofloxacin, danofloxacin, difloxacin, enrofloxacin, gatifloxacin, levofloxacin, marbofloxacin, and orbifloxacin.
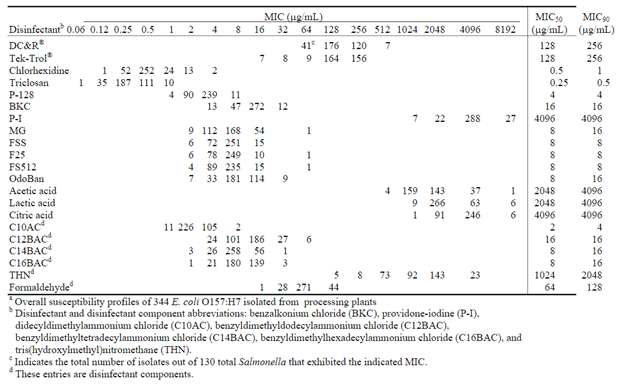
Disinfectant resistance was determined by broth microdilution according to CLSI standards for antimicrobial susceptibility testing. The disinfectants tested were chlorhexidine diacetate, DC&R®, Tek-Trol®, P-128, Betadine® solution, formaldehyde, benzalkonium chloride, triclosan, MG, F25, FSS, F512, OdoBan, and the disinfectant components didecyldimethylammonium chloride (C10AC), benzyldimethyldodecylammonium chloride (C12BAC), benzyldimethyltetradecylammonium chloride (C14BAC), benzyldimethylhexadecylammonium chloride (C16BAC), and tris(hydroxylmethyl)nitromethane (THN). Sterile dimethyl sulfoxide (DMSO) was used to solubilize some disinfectants, and reverse osmosis water (H2O) was produced on site by a reverse osmosis system obtained from Millipore Corp.
Pulse-field gel electrophoresis (PFGE) analysis was performed according to the protocol developed by the Centers for Disease Control and Prevention (CDC), and the banding patterns were analyzed using Bionumerics software.
Polymerase chain reaction (PCR) analysis for the disinfectant sugE gene used primers: forward 5’-ATCGGGTTAGCGGACTCAC-3’ and reverse 5’-ATTGCTGGTCTGCTGGAAGT-3’, and the PCR-replicon typing of E. coli O157:H7 strains was performed.
Findings
The disinfectant and antibiotic susceptibility profiles of 344 E. coli O157:H7 cattle feces, hide, carcass, and ground meat isolates from different parts of the United States were determined. A low incidence of antibiotic resistance was observed (14%, Table 1). The highest incidence of resistance was observed to FIS (10.5%), TET (9.9%), STR (7%), and CHL (4.9%). Two ground meat, one cull cow hide and previseration isolate were resistant to 8 antibiotics. Table 2 shows the overall disinfectant MICs. There were 69 (20%) of the isolates resistant to chlorhexidine, benzalkonium chloride, or had elevated MICs to benzyldimethyldodecylammonium chloride. All isolates were pansusceptible to triclosan. All 344 isolates were positive for the sugE gene, and sugE did not appear to correlate with the quarternary ammonium chlorides tested. There was spontaneous resistance (SR) at elevated levels of antibiotics (1.4%) and disinfectants (6.1%). The highest rate of SR was observed in OdoBan, ammonium chloride components, and the surface disinfectants F25, FS512, and MG used in dairies, restaurants, and food processing plants. SR could allow the presence of bacteria where they should have been removed by disinfection. High MICs (1024–4096 μg/mL) were found for the acids, acetic, lactic, and citric. The decreasing order of acid potency based on MICsmolar was acetic, citric, and lactic acid. PFGE analysis was performed using XbaI digests of the E. coli O157:H7, and clusters 1A, 1B, 1C, and 1H were associated with high multi-drug resistance (MDR), but little disinfectant resistance. PFGE cluster 1C was associated with resistance to 8 antibiotics in two ground meat isolates. Similar resistance traits were observed in a cull cow hide and previseration isolate with PFGE cluster designation 1H. There was no correlation between disinfectant resistance and antibiotic resistance in the E. coli O157:H7 isolates tested.
Implications
Cattle hides did not appear to be a source of MDR, but a low incidence of MDR was observed in isolates from feedlot fecal samples as well as from cull cows. Some of the highest levels of E. coli O157:H7 MDR (8 different antibiotics) were found in ground meat samples, which resembled antibiotic resistance traits observed from isolates from cull cows, and may likely be “outbreak” strains. Perhaps cull cows may be an important source of E. coli O157:H7 MDR, and in that regard, the application of chlorate prior to shipment may be an important way to reduce levels of MDR in cull cows.
The highest incidence of resistance to benzalkonium chloride was also observed in cull cows. A low incidence of chlorhexidine resistance was observed from cattle hides, cull cows, and from feedlots; however, chlorhexidine resistance was most often associated with ground meat samples. Ground meat isolates also showed higher MICs for C12 benzylammonium chloride. Perhaps the higher MIC levels to disinfectants in ground meat isolates results from using ammonium chlorides in and around production facilities. Spontaneous resistance was observed in 6.1% of the 344 isolates tested at elevated levels of various disinfectants, predominately those containing quaternary ammonium chlorides. The spontaneous resistance at elevated disinfectant concentrations could be problematic by allowing some bacteria not to be neutralized by a disinfectant application. This could allow the presence of bacteria where they should have been removed in dairies, restaurants, or food processing plants.
The study confirmed that E. coli O157:H7 demonstrated high MICs with the three acids, acetic, lactic, and citric, which is consistent with the acid resistance of E. coli O157:H7. The decreasing order of acid potency was acetic, citric, and lactic acid. Overall, there did not appear to be a correlation between disinfectant resistance and antibiotic resistance in the E. coli O157:H7 tested.
Table 1. NARMS overall antibiotic MICs and resistance profiles among E. coli isolatesa
|
MIC50 Antibioticb (mg/mL) |
MIC90 (mg/mL) |
Range (mg/L) |
No. (%) Resistant |
Break Point |
|
AMI 2 |
2 |
£ 0.5 – 8 |
0 (0) |
³64 |
|
AMP 2 |
4 |
£ 1 – >32 |
9 (2.6) |
³32 |
|
AUG 4/2 |
4/2 |
£ 1/0.5 – >32/16 |
4 (1.2) |
³32/16 |
|
AXO £ 0.25 |
£ 0.25 |
£ 0.25 – 32 |
0 (0) |
³64 |
|
CHL 8 |
16 |
4 – >32 |
17 (4.9) |
³32 |
|
CIP 0.03£ 16 FIS |
0.03>256 |
£ 0.015 – 0.25 £ 16 – >256 |
0 (0) – (–)c |
³4³512 |
|
FOX 8 |
8 |
2 – >32 |
4 (1.2) |
³32 |
|
GEN 0.5 |
1 |
£ 0.25 – 8 |
0 (0) |
³16 |
|
KAN £ 8 |
£ 8 |
£ 8 – >64 |
4 (1.2) |
³64 |
|
NAL 4 |
4 |
1 – >32 |
1 (0.3) |
³32 |
|
STR £ 32 |
£ 32 |
£ 32 – >64 |
24 (7.0) |
³64 |
|
SXT £ 0.12/2.38 |
£ 0.12/2.38 |
£ 0.12/2.38 – ³4/76 |
2 (0.6) |
³4/76 |
|
TET £ 4 |
8 |
£ 4 – >32 |
34 (9.9) |
³16 |
|
XNL 0.5 |
1 |
0.25 – >8 |
8 (2.3) |
³8 |
ANARMS Gram-negative antibiotic overall MIC profiles of 344 E. coli O157:H7 isolates.
BAntibiotic abbreviations: Amikacin (AMI), Ampicillin (AMP), Amoxicillin/Clavulanic acid (AUG), Ceftriazone (AXO), Chloramphenicol (CHL), Ciprofloxacin (CIP) , Sulfisoxazole (FIS), Cefoxitin (FOX), Gentamicin (GEN), Kanamycin (KAN), Nalidixic acid (NAL), Streptomycin (STR), Trimethoprim/Sulfamethoxazole (SXT), Tetracycline (TET), and Ceftiofur (XNL).
CThe FIS range on the antibiotic plates is not large-enough to observe the resistance break point.
Table 2. Distribution of disinfectant and disinfectant component susceptibility profiles among E. coli O157:H7