Project Summary
Animal-Level Predictors of Multiple Drug-Resistant Salmonella Carriage by Cull Dairy Cattle
- Principle Investigator(s):
- Guy H. Loneragan1, Mindy M. Brashears2, Rebecca M. McCarthy1, David Nisbet3, Tom Edrington3, Thomas E. Besser4, and Angela Daniels5
- Institution(s):
- 1West Texas A&M University
2Texas Tech University
3USDA-ARS
4Washington State University
5Circle H Animal Health LLC - Completion Date:
- 2009
Background
Salmonella is an important cause of food-borne illness in the US and beef has been identified as a source. Culled dairy cows are a significant source of beef products, particularly ground beef, and they also frequently carry Salmonella. Understanding factors that contribute to Salmonella carriage in dairy cattle is important for control purposes.
This study was undertaken to estimate herd-level burden and identify animal-level predictors of Salmonella carriage in dairy cattle of the southern High Plains.
Methodology
Fecal samples were collected from cull cows either just prior to shipment from the dairy or at a regional auction market. Samples were cultured for Salmonella using various media and where possible, 3 distinct colonies per sample were collected. For samples from which Salmonella was recovered, the researchers attempted to quantify the concentration of Salmonella in the feces. In addition to cull cows, a selection of milking cows on 3 dairies was enrolled and feces were collected; in addition to collecting feces, a 1000-cm2 area of hide over the top of the animal was swabbed. Various methods were used to culture Salmonella from these samples. Antibiotic susceptibility and serotypes were determined.
Findings
Feces were collected from 706 cows culled from 9 dairies. Salmonella was recovered from 32.6% of cull cattle and varied by month and dairy.
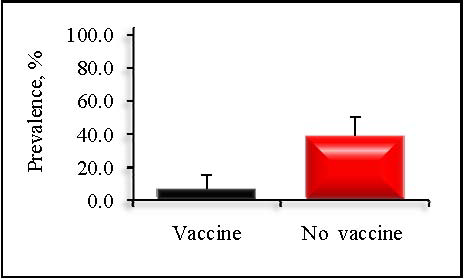
Prevalence at the dairy level varied from 4.4 to 86.3% while sample-level concentration varied from -0.5 to 7.0 log10 MPN per g feces. Dairies that administered a vaccine containing siderophore receptors and porin proteins (SRP) were associated with a significantly lower prevalence of Salmonella than herds that did not vaccinate (7.6 vs. 39.2%; P < 0.01; Figure 1).
In addition, 70 paired fecal and hide samples were collected from milking cows housed on 3 dairies; Salmonella was recovered from 41.3% of cattle (34.3% of hides and 30.0% of fecal samples). Dairy-level recovery varied from 10 to 100%. One, two and three serovars were recovered from 62.5, 18.8, and 18.7% of positive fecal samples from milking cows, respectively; 1, 2, 3, and 4 serovars were recovered from 16.7, 20.8, 41.7, and 20.8% of hides, respectively. Of all isolates (cull and milking cows), the vast majority (>90%) were pansusceptible or resistant to 1 drug and were of serovars not typically observed in human or animal diseases. Of the isolates from cull cows, 3.6% were resistant to more than 4 drugs (all but 1 were Salmonella Newport and all but 1 were recovered from just 1 of the 9 dairies).
Implications
Salmonella is commonly recovered from dairy cattle in the Texas High Plains but the majority of salmonellae appear to be non-pathogenic commensals. Moreover, many cattle harbor multiple serovars. While not a primary objective of this study, vaccination with Salmonella-specific SRP was associated with reduced prevalence and warrants further investigation. Because most variation occurs at the herd level, the most appropriate approach to systematically study this vaccine is a multi-herd study where whole herds are either administered the vaccine or not.