Project Summary
Survey of the Prevalence of Escherichia coli O157:H7 on the Surface of Sub-Primal Cuts of Beef During Winter Months (Phase I)
- Principle Investigator(s):
- J. E. (Ken) Kennedy
- Institution(s):
- ABC Research Corporation
- Completion Date:
- May 2004
Background
The USDA FSIS published a notice in the Federal Register of January 19, 1999 stating its intention to include non-intact beef products in the Escherichia coli O157:H7 testing program. Concerns that prompted the release of this notice were that intact cuts of beef further processed into non-intact products may have pathogens introduced below the meat surface and therefore require a higher internal cook temperature to render the product safe for consumption.
In June 2003, a Chicago processor of steaks and other meat products, voluntarily recalled 739,000 pounds of vacuum packaged steaks and other meat products injected with meat tenderizers due to possible E. coli O157:H7 contamination. This recent recall highlights the need determine the extent to which E. coli O157:H7 is present on the surface of sub-primal cuts prior to mechanical tenderization.
The stated objective for this work was to determine the prevalence of E. coli O157:H7 and indicator microorganisms on the surface of beef sub-primals prior to the enhancement process. This data was collected during the winter months.
Methodology
PRODUCTS
Six representative sub-primal cut types representing 3 commodity and 3 closely trimmed cuts were sampled in this study. Products included “chuck tenders”, “1/4 trimmed strips”, “bottom round flat”, “rough trimmed brisket”, “cap-on top round” and “cap-off insides”. One hundred samples each of the six sub-primal types were collected from five plants (i.e., total of 600 samples) located in the Midwest, the southern Midwest, the northern Midwest and the Southeast over a six-week period between 1/6/04 and 2/19/04.
SAMPLE COLLECTION AND SHIPPING
Samples were collected from the entire surface of each sub-primal just prior to the beef enhancement process using the sponge technique. Sponges were hydrated in 20 mL Buffered Peptone water (BPW). An exception to this technique occurred with Plant E in that their sponges were hydrated in 10 rather than 20 mL BPW. Each sub-primal was sampled using only one sponge. Both sides of the sponge were used to sample each sub-primal. Each individual sub-primal bag was considered a lot and placed on “hold and test” status until results for E. coli O157:H7 analyses were completed. Product was handled and held differently depending on the product and facility.
The sponge samples were collected at the respective plants of the industry cooperator/collaborators, aliquots were removed for indicator organism analyses, and the sponge samples were shipped on the same day to ABC Research Corp. unless they were collected during second shift. In that case, samples were sent the following day. Sponge samples (in stomacher bags) were shipped in insulated containers with ice packs in a way to assure that sponges were not frozen but remained at refrigeration temperature during transport via overnight courier. Each sample shipment included the following information: the lot, the product code and age of the product. A sample analysis request form was completed for each set of samples sent to the ABC Research Corp. Upon receipt at the laboratory, the temperature of each cooler was taken and documented in the laboratory notebook.
MICROBIOLOGICAL ANALYSES
Indicator organism analyses (conducted on-site by industry cooperator/collaborators).
Sponge samples were stomached for 60 seconds to ensure sample homogeneity. Two (2) mL of the sponge diluent/rinsate was removed for Aerobic Plate Counts (APC) and generic E. coli/coliform analyses using 3M Petrifilm™). The 2 mL aliquot was considered the “0” dilution” and only the “0” dilution was plated for APC and E. coli/coliform counts. APC plates were incubated at 35°C for 48 h and enumerated using standard counting procedures. E. coli/coliforms plates were incubated at 35°C for 24 h and enumerated using standard counting procedures.
E. coli O157:H7 Analyses (conducted by ABC Research Corp.)
Sponge samples were stomached for 60 seconds to ensure sample homogeneity. A 1 mL aliquot from the sponge sample was extracted for use in the most probable number procedure (MPN) procedure developed at USDA Meat Animal Research Center.
For the E. coli O157:H7 presence/absence analyses, each sponge (and associated rinsate) was enriched with 80 mL of trypticase soy broth (TSB). The TSB was tempered to 25°C prior to addition to sponges. After addition of the TSB, the sponge and TSB was stomached for 60 seconds on medium speed. Prior to incubation, sponge samples were checked to make certain sponges are turned so they are immersed in enrichment media. A positive and a negative control sample were included in each set of analyses. Samples and controls were incubated for 2 hours at 25°C and subsequently for 10 additional hours at 42°C. After the 12-hour incubation, samples were held overnight at 4°C. Samples were then analyzed for the presence of E. coli O157:H7 using BAX-PCR methodology. Samples positive for the presence of E. coli O157:H7 were enumerated using the most probable number (MPN) procedure.
For the MPN sample preparation, deep-well microtiter plates with 900 μL of TSB in each well were prepared as shown in Table A (see below). Each sample was analyzed using a 3 well x 4 dilutions MPN series each made from the original 3 wells as follows:
- The 1.0 mL sample aliquot tube was vortexed and 100 μL added to each of three consecutive wells in the microtiter plate.
- The first three wells were inoculated for each of the 8 samples. Utilizing an 8 or 12 channel micropipettor, samples were thoroughly mixed. After the third aspiration, 100 μL was transferred from the first row to the second row. After completion of the first transfer, pipette tips were replaced and the aspiration and transfer repeated for remaining rows, until all dilutions were made for the entire plate.
- Once all dilutions were made, plates were labeled and covered with microtiter well covers. All plates were held at room temperature until all samples were prepared for incubation. The inoculated MPN trays were incubated at 25°C for 2 hours and subsequently at 42°C for an additional 10 hours. Microtiter trays were then held at 4°C until BAX results were obtained.
- For samples that tested negative for E. coli O157:H7 using the BAX-PCR methodology, the MPN procedure was discontinued and the microtiter plates discarded.
- For samples testing positive for E. coli O157:H7, the MPN analysis was initiated.
- For positive samples, all twelve-microtiter plate MPN wells were processed using the BAX-PCR protocol. MPN counts (per mL of sponge diluent/rinsate) were derived from FDA/BAM MPN tables. Those MPN results were then used to calculate the most probable number of E. coli O157:H7 per original sub-primal sample. It should be noted that no microtiter plates were further processed via BAX-PCR because no E. coli O157:H7 positives samples were observed in the study.
Findings
E. coli O157:H7.
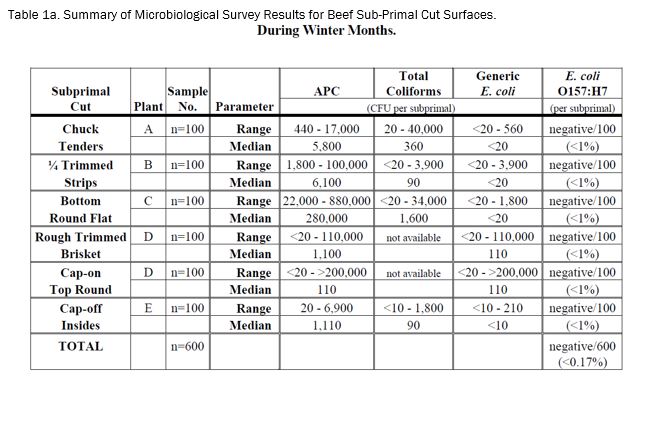
The results of the E. coli O157:H7 survey of six representative types (100 samples each) of beef sub-primal cuts intended for mechanical tenderization (enhancement) during the winter months (i.e., January and February 2004) are presented in Table 1a. No E. coli O157:H7 was detected on the surface of any of the 600 beef sub-primal samples analyzed over the six-week survey period (i.e., 1/6/04 - 2/19/04). Thus, the overall incidence of E. coli O157:H7 was less than 0.17%.
Indicator microorganisms.
The results of the corresponding indicator organism survey of the six types of beef sub-primal cuts during the winter months are also presented in Table 1a. The recovery of aerobic plate counts, total coliforms and generic E. coli were variable within each sample set of each sub-primal and between sub-primal types. It should be noted that the levels of generic E. coli were very low and/or undetectable for most of the sub-primal samples. In fact, generic E. coli was not detected in 81, 78, 94 and 83%, respectively, of chuck tenders, trimmed strips, bottom round flat, and cap-off insides samples.
Implications
These results indicate that the incidence of E. coli O157:H7 contamination on the surface of beef sub-primal cuts intended for mechanical tenderization (enhancement) is very low (i.e., <0.17%) with no E. coli O157:H7 being detected in any of the 600 sub-primal samples during January and February of 2004.
A previous study demonstrated an extremely low internalization rate of surface contaminating E. coli O157:H7 into subprimals by blade tenderization. These results along with those of the previous internalization study indicate that internal contamination of beef sub-primals with E. coli O157:H7 via mechanical tenderization (enhancement) is a very improbable phenomenon.