Project Summary
Phage-Encoded Dissemination of Shiga Toxin Genes Among Bovine E. coli
- Principle Investigator(s):
- Jeffery LeJeune
- Institution(s):
- Food Animal Health Research Program, The Ohio State University
- Completion Date:
- May 2006
Background
Although Escherichia coli O157:H7 has been the most prevalent Shiga toxin producing serotype of Escherichia coli (STEC) in the United States, more than 900 isolates of non-O157 STEC, including serogroups O26, O111, O121, O45 and O145, were cultured from people with sporadic illnesses from 43 different states between 1983 and 2002. In all countries outside North America, non-O157 serogroups of Shiga toxin-producing E. coli (STEC) have emerged as the cause about four times as many cases of human disease as E. coli O157:H7. It is estimated that non-O157 STEC account for as much as half of all human STEC infections in the United States and Canada.
Since non-O157 infections are probably under-diagnosed and sporadic in nature, the reservoirs of these organisms are poorly understood. Studies in other countries have shown that cattle may harbor non-O157 STEC. However, there is very limited information concerning the prevalence of non-O157 STEC in U.S. cattle. There are at least two independent routes by which pathogenic STEC can end up in cattle. First, dissemination of bacteria from a non-bovine source (e.g. environmental exposure) may result in colonization of cattle. In fact, two human outbreaks of non-O157 have been linked to environmental exposure.
Secondly, it is considered possible that new strains of Shiga toxin-producing E. coli may emerge by acquiring the ability to produce Shiga toxin from E. coli O157 following the exchange of genetic material. This hypothesis is supported by laboratory experiments that have demonstrated the potential for bacteriophages from E. coli O157 that encode Shiga toxins to lysogenize new host bacteria, making them STEC.
The extent to which these two mechanisms contribute to the emergence of non-O157 STEC in U.S. cattle populations is important to the beef industry. Understanding how non-O157 STEC emerges in cattle populations should provide valuable insight into the control and prevention of this pathogen.
The objectives of this study were as follows:
- Determine and compare the diversity of stx2-converting phages isolated from bovine STEC of US and foreign origin.
- Define the host range and determine the frequency of lysogenization of E. coli of US bovine origin by stx2-converting phages from US E. coli O157:H7.
- Determine the efficacy of lysogenization of non-O157 bacteria host cells with O157-origin stx2-converting phages within live cattle (in situ).
Genetic Diversity
Genes present in the stx2-encoding phage of E. coli 933W and in phage lambda were identified through GenBank searches. Primers designed to target these genes were then used to screen a panel of E. coli O157 and E. coli O26 isolates acquired from diverse geographic locations.
Host Range
The Shiga toxin gene was replaced with an antibiotic marker (chloramphenicol acetyl transferase, CAT) gene among isolates of E. coli O157 and isolates of E. coli O26. The ability of these genetically-tagged phages to infect (lytic and lysogenic) E. coli O157, E. coli O26 and a panel of generic E. coli isolated from cattle was assayed by the drop plate method and recovery of chloramphenicol encoding lysogens.
in vivo Lysogenization of stx-encoding Phages
Eight, 12-week-old wean calves were inoculated with an E. coli O157 strain in which the Shiga toxin gene had been replaced with a kanamycin resistance gene (E. coli O157 stxÄkan). The magnitude of the inoculum strain, as well as other kanamycin-resistant, Gram-negative flora in the calves was followed. Kanamycinpositive, sorbitol-positive (non-O157) isolates were screened by PCR for stx-phage sequences.
A CAT-encoding E. coli O157 stx-knock out engineered in the host-range study was used to quantitatively assess the in vitro impact of induction by several commonly used therapeutic antibiotics (Micotil, Nuflor, Baytril, Naxcel, and Tetracycline). The magnitude of chloramphenicol acetyl transferase production from the E. coli O157 Ä CAT isolate was measured under induction in the presence and absence of bile salts.
Findings
Diversity of Phages
Previously the genetic determinates immediately upstream of the stx2 gene in over 100 E. coli O157 isolates had been determined, including isolates from Japan, Scotland, Australia, and the United States. All of these isolates tested positive for either antiterminator Q gene present in phage 933W or phage 21. In addition, a panel of 42 isolates for other phage-related genome sequence was tested, including isolates of E. coli O111, O26 and O157. None of the non-O157 isolates tested positive for either of the two Q alleles present in the O157 isolates.
Host range
Attempts to infect under laboratory conditions the phages derived from the E. coli O157 Ä CAT isolates created above were unsuccessful. The O157 and O26 phages neither lysed nor lysogenize other O157, O26 or generic E. coli isolated from cattle or the environment.
in vivo Lysogenization
No lysogens were obtained from any of the four calves challenged with the kanamycin-encoding E. coli O157 strain. There was some background E. coli flora present in the calves that were resistant to kanamycin, however screening of these colonies by PCR demonstrated that these colonies did not have the stx-endocing phage sequences around the kanamycin gene, therefore they were not lysogens. Given the high level of E. coli O157 in these cattle, the frequency and volume of samples analyzed, and the vast variety of potential host bacteria, if lysogenization of stx-encoding phages occurs at all in live cattle, the frequency must be rare.
Previous studies demonstrated that some growth-promoting antimicrobials fed to cattle were able to greatly induce the amount of stx-phage released by E. coli when the bacteria were exposed to these agents in vitro. Testing has begun for several commonly used therapeutic antibiotics (Micotil, Nuflor, Baytril, Naxcel, and Tetracycline) on their ability to induce phage in vitro.
Similar results were observed regardless of whether or not bile salts were included in the test medium. Although preliminary, results suggest that at minimum inhibitory concentration of each antibiotic, these agents induce phage release marginally, if at all (See Figure 1).
In summary, the diversity of bacteriophages that encode the stx2 gene among STEC is great. Some of this diversity may provide protection from stx-enducing phages (difference in host range) whereas common genes such as cro and cI may play a role in immunity to superinfection should these phages manage to infect new host cells. The evidence from the epidemiological and the in vivo experiments conducted in this study are inconsistent with the hypothesis of non-O157 emergence from endogenous bovine sources in the U.S.
IMPLICATIONS
The results of this study strongly suggest that current livestock management practices do not promote the emergence of new strains of pathogenic E. coli from the current pool of E. coli O157 in U.S. livestock populations. Instead, preventing non-O157 E. coli in U.S. cattle will rely on prevention of highly pathogenic non-O157 from being introduced to U.S. cattle from outside sources.
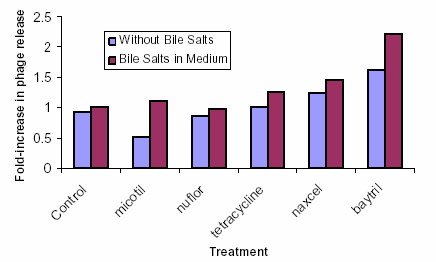
Figure 1: Effects of Therapeutic Antibiotics at Minimal Inhibitory Concentrations on Induction of stx2-Encoding Phages